Crystal structure of Tcur_1030 protein from Thermomonospora curvata
Michalska, K., Li, H., Endres, M., Joachimiak, A.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Uncharacterized protein | 151 | Thermomonospora curvata DSM 43183 | Mutation(s): 0 Gene Names: Tcur_1030 | 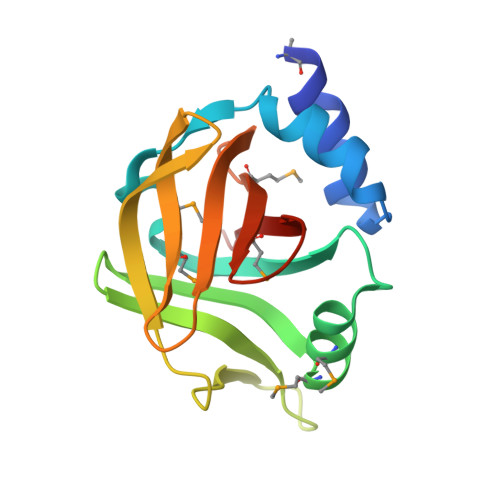 | |
UniProt | |||||
Find proteins for D1A7M7 (Thermomonospora curvata (strain ATCC 19995 / DSM 43183 / JCM 3096 / KCTC 9072 / NBRC 15933 / NCIMB 10081 / Henssen B9)) Explore D1A7M7 Go to UniProtKB: D1A7M7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | D1A7M7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| MSE Query on MSE | A, B | L-PEPTIDE LINKING | C5 H11 N O2 Se |  | MET |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 66.803 | α = 90 |
| b = 66.803 | β = 90 |
| c = 127.368 | γ = 120 |
| Software Name | Purpose |
|---|---|
| SBC-Collect | data collection |
| SHELX | model building |
| MLPHARE | phasing |
| DM | model building |
| ARP/wARP | model building |
| Coot | model building |
| BUSTER | refinement |
| HKL-3000 | data reduction |
| HKL-3000 | data scaling |
| SHELX | phasing |
| DM | phasing |