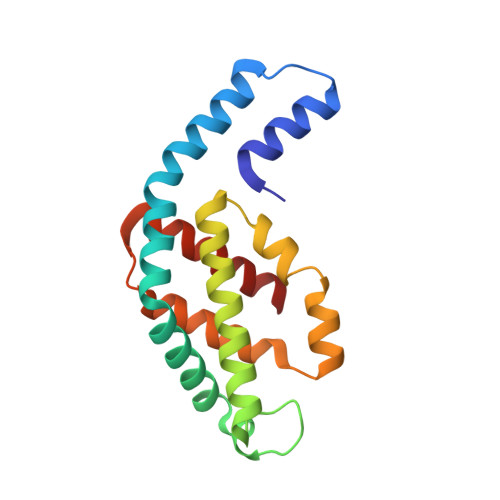
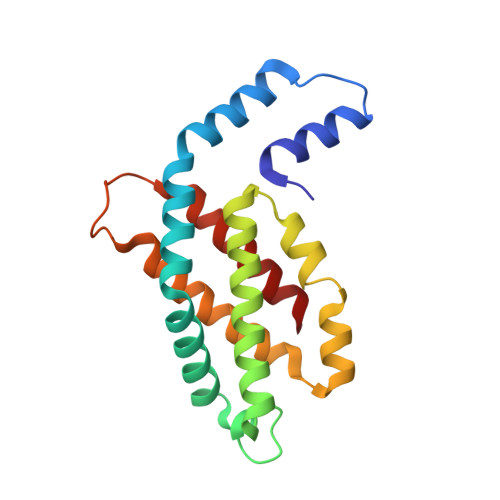
Crystal Structure and Interaction of Phycocyanin with beta-Secretase: A Putative Therapy for Alzheimer's Disease.
Singh, N.K., Hasan, S.S., Kumar, J., Raj, I., Pathan, A.A., Parmar, A., Shakil, S., Gourinath, S., Madamwar, D.(2014) CNS Neurol Disord Drug Targets 13: 691-698
- PubMed: 24576002
- DOI: https://doi.org/10.2174/1871527313666140228114456
- Primary Citation of Related Structures:
4L1E - PubMed Abstract:
Alzheimer's disease (AD) represents a neurological disorder, which is caused by enzymatic degradation of an amyloid precursor protein into short peptide fragments that undergo association to form insoluble plaques. Preliminary studies suggest that cyanobacterial extracts, especially the light-harvesting protein phycocyanin, may provide a means to control the progression of the disease. However, the molecular mechanism of disease control remains elusive. In the present study, intact hexameric phycocyanin was isolated and crystallized from the cyanobacterium Leptolyngbya sp. N62DM, and the structure was solved to a resolution of 2.6 A. Molecular docking studies show that the phycocyanin αβ-dimer interacts with the enzyme β-secretase, which catalyzes the proteolysis of the amyloid precursor protein to form plaques. The molecular docking studies suggest that the interaction between phycocyanin and β-secretase is energetically more favorable than previously reported inhibitor-β-secretase interactions. Transgenic Caenorhabditis elegans worms, with a genotype to serve as an AD-model, were significantly protected by phycocyanin. Therefore, the present study provides a novel structure-based molecular mechanism of phycocyanin-mediated therapy against AD.
Organizational Affiliation:
(Datta Madamwar) BRD School of Biosciences, Sardar Patel Maidan, Vadtal Road, Satellite Campus, Post Box No. 39, Sardar Patel University, Vallabh Vidyanagar-388 120, Gujarat, India. datta_madamwar@yahoo.com.