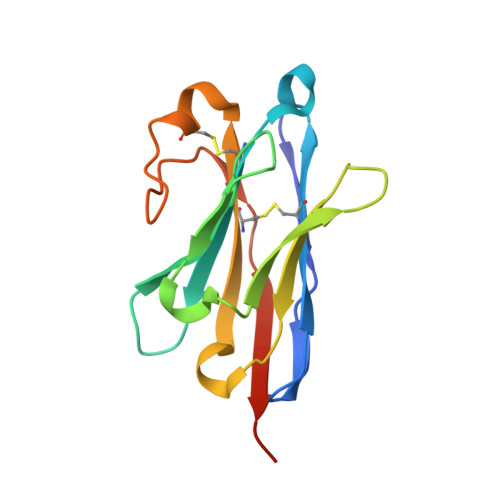
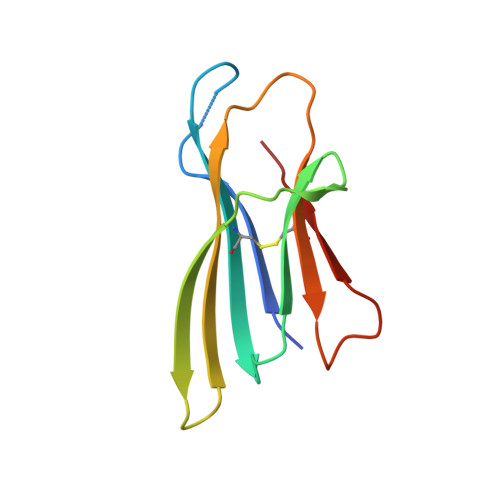
Structure of an early native-like intermediate of beta 2-microglobulin amyloidogenesis.
Vanderhaegen, S., Fislage, M., Domanska, K., Versees, W., Pardon, E., Bellotti, V., Steyaert, J.(2013) Protein Sci 22: 1349-1357
- PubMed: 23904325
- DOI: https://doi.org/10.1002/pro.2321
- Primary Citation of Related Structures:
4KDT - PubMed Abstract:
To investigate early intermediates of β2-microglobulin (β2m) amyloidogenesis, we solved the structure of β2m containing the amyloidogenic Pro32Gly mutation by X-ray crystallography. One nanobody (Nb24) that efficiently blocks fibril elongation was used as a chaperone to co-crystallize the Pro32Gly β2m monomer under physiological conditions. The complex of P32G β2m with Nb24 reveals a trans peptide bond at position 32 of this amyloidogenic variant, whereas Pro32 adopts the cis conformation in the wild-type monomer, indicating that the cis to trans isomerization at Pro32 plays a critical role in the early onset of β2m amyloid formation.
Organizational Affiliation:
Structural Biology Research Centre, VIB, Pleinlaan 2, 1050, Brussel, Belgium; Structural Biology Brussels, Vrije Universiteit Brussel, Pleinlaan 2, 1050, Brussel, Belgium.