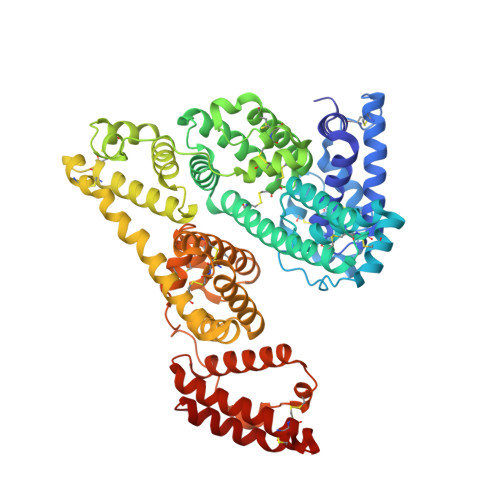
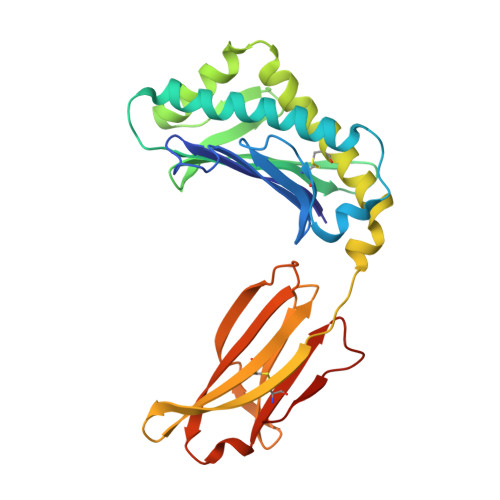
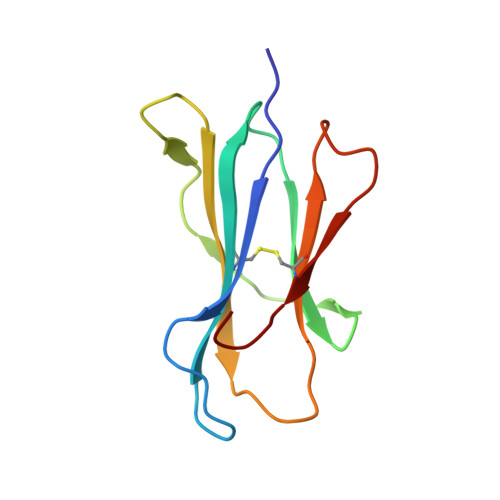
Crystal structure of an HSA/FcRn complex reveals recycling by competitive mimicry of HSA ligands at a pH-dependent hydrophobic interface.
Schmidt, M.M., Townson, S.A., Andreucci, A.J., King, B.M., Schirmer, E.B., Murillo, A.J., Dombrowski, C., Tisdale, A.W., Lowden, P.A., Masci, A.L., Kovalchin, J.T., Erbe, D.V., Wittrup, K.D., Furfine, E.S., Barnes, T.M.(2013) Structure 21: 1966-1978
- PubMed: 24120761
- DOI: https://doi.org/10.1016/j.str.2013.08.022
- Primary Citation of Related Structures:
4K71 - PubMed Abstract:
The long circulating half-life of serum albumin, the most abundant protein in mammalian plasma, derives from pH-dependent endosomal salvage from degradation, mediated by the neonatal Fc receptor (FcRn). Using yeast display, we identified human serum albumin (HSA) variants with increased affinity for human FcRn at endosomal pH, enabling us to solve the crystal structure of a variant HSA/FcRn complex. We find an extensive, primarily hydrophobic interface stabilized by hydrogen-bonding networks involving protonated histidines internal to each protein. The interface features two key FcRn tryptophan side chains inserting into deep hydrophobic pockets on HSA that overlap albumin ligand binding sites. We find that fatty acids (FAs) compete with FcRn, revealing a clash between ligand binding and recycling, and that our high-affinity HSA variants have significantly increased circulating half-lives in mice and monkeys. These observations open the way for the creation of biotherapeutics with significantly improved pharmacokinetics.
Organizational Affiliation:
Eleven Biotherapeutics, Inc., Cambridge, MA 02142, USA.