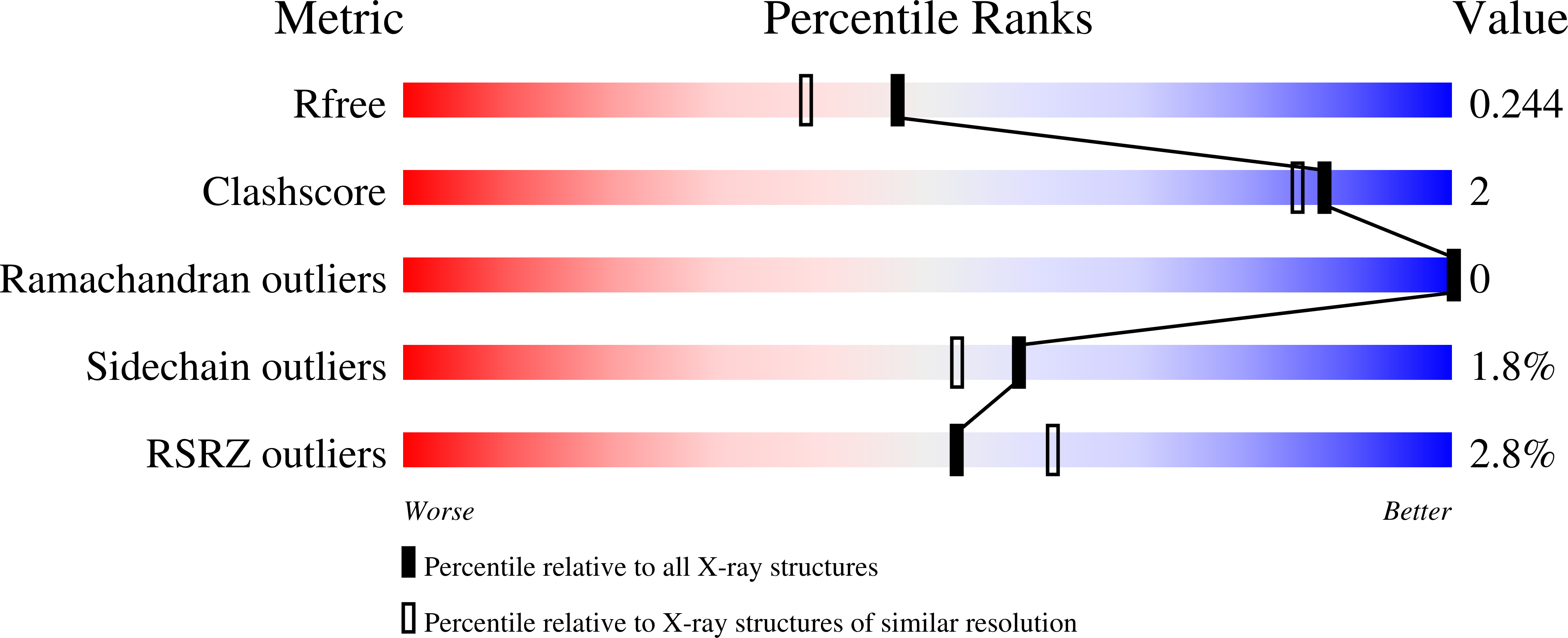
Development of potent and selective Plasmodium falciparum calcium-dependent protein kinase 4 (PfCDPK4) inhibitors that block the transmission of malaria to mosquitoes.
Vidadala, R.S., Ojo, K.K., Johnson, S.M., Zhang, Z., Leonard, S.E., Mitra, A., Choi, R., Reid, M.C., Keyloun, K.R., Fox, A.M., Kennedy, M., Silver-Brace, T., Hume, J.C., Kappe, S., Verlinde, C.L., Fan, E., Merritt, E.A., Van Voorhis, W.C., Maly, D.J.(2014) Eur J Med Chem 74: 562-573
- PubMed: 24531197
- DOI: https://doi.org/10.1016/j.ejmech.2013.12.048
- Primary Citation of Related Structures:
4JBV - PubMed Abstract:
Malaria remains a major health concern for a large percentage of the world's population. While great strides have been made in reducing mortality due to malaria, new strategies and therapies are still needed. Therapies that are capable of blocking the transmission of Plasmodium parasites are particularly attractive, but only primaquine accomplishes this, and toxicity issues hamper its widespread use. In this study, we describe a series of pyrazolopyrimidine- and imidazopyrazine-based compounds that are potent inhibitors of PfCDPK4, which is a calcium-activated Plasmodium protein kinase that is essential for exflagellation of male gametocytes. Thus, PfCDPK4 is essential for the sexual development of Plasmodium parasites and their ability to infect mosquitoes. We demonstrate that two structural features in the ATP-binding site of PfCDPK4 can be exploited in order to obtain potent and selective inhibitors of this enzyme. Furthermore, we demonstrate that pyrazolopyrimidine-based inhibitors that are potent inhibitors of the in vitro activity of PfCDPK4 are also able to block Plasmodium falciparum exflagellation with no observable toxicity to human cells. This medicinal chemistry effort serves as a valuable starting point in the development of safe, transmission-blocking agents for the control of malaria.
Organizational Affiliation:
Department of Chemistry, University of Washington, Seattle, WA 98195-1700, USA.