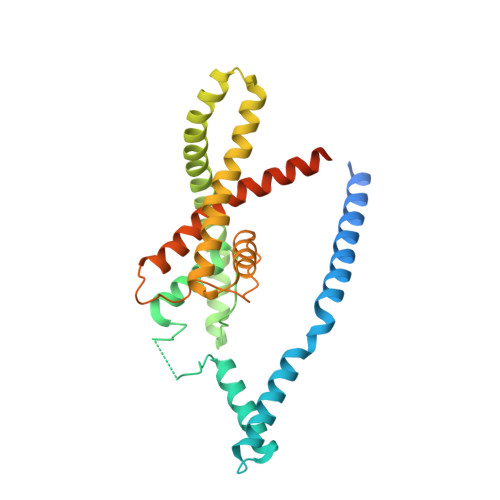
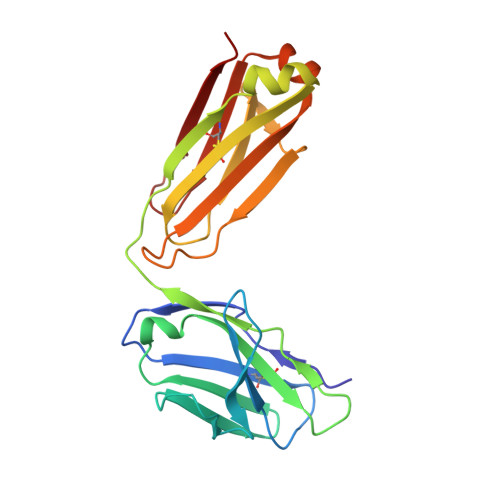
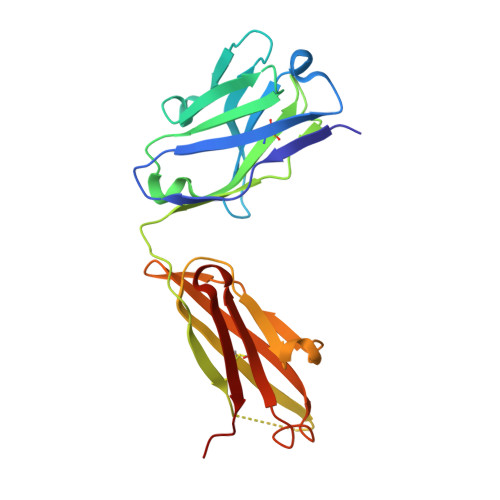
Domain-swapped chain connectivity and gated membrane access in a Fab-mediated crystal of the human TRAAK K+ channel.
Brohawn, S.G., Campbell, E.B., Mackinnon, R.(2013) Proc Natl Acad Sci U S A 110: 2129-2134
- PubMed: 23341632
- DOI: https://doi.org/10.1073/pnas.1218950110
- Primary Citation of Related Structures:
4I9W - PubMed Abstract:
TRAAK (TWIK-related arachidonic acid-stimulated K(+) channel, K2P4.1) K(+) ion channels are expressed predominantly in the nervous system to control cellular resting membrane potential and are regulated by mechanical and chemical properties of the lipid membrane. TRAAK channels are twofold symmetric, which precludes a direct extension of gating mechanisms that close canonical fourfold symmetric K(+) channels. We present the crystal structure of human TRAAK in complex with antibody antigen-binding fragments (Fabs) at 2.75-Å resolution. In contrast to a previous structure, this structure reveals a domain-swapped chain connectivity enabled by the helical cap that exchanges two opposing outer helices 180° around the channel. An unrelated conformational change of an inner helix seals a side opening to the membrane bilayer and is associated with structural changes around the K(+)-selectivity filter that may have implications for mechanosensitivity and gating of TRAAK channels.
Organizational Affiliation:
Laboratory of Molecular Neurobiology and Biophysics and Howard Hughes Medical Institute, The Rockefeller University, New York, NY 10065, USA.