The crystal structure of a short-chain dehydrogenases/reductase (wide type) from Veillonella parvula DSM 2008 in complex with NADP.
Tan, K., Hatzos-Skintges, C., Clancy, S., Joachimiak, A.To be published.
Experimental Data Snapshot
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| NAD-dependent epimerase/dehydratase | 221 | Veillonella parvula DSM 2008 | Mutation(s): 0 Gene Names: Vpar_0111 | 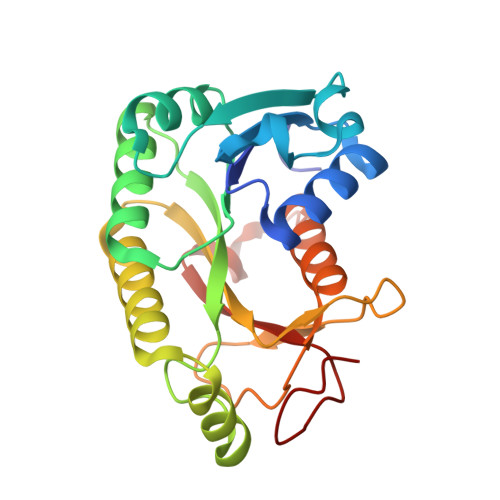 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 5 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| NAP Query on NAP | C [auth A], K [auth B] | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE C21 H28 N7 O17 P3 XJLXINKUBYWONI-NNYOXOHSSA-N |  | ||
| TEA Query on TEA | J [auth B] | TRIETHYLAMMONIUM ION C6 H16 N ZMANZCXQSJIPKH-UHFFFAOYSA-O |  | ||
| GOL Query on GOL | E [auth A], F [auth A], G [auth A] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| FMT Query on FMT | H [auth A], I [auth A], M [auth B] | FORMIC ACID C H2 O2 BDAGIHXWWSANSR-UHFFFAOYSA-N |  | ||
| CL Query on CL | D [auth A], L [auth B] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 109.219 | α = 90 |
| b = 71.687 | β = 121.44 |
| c = 65.016 | γ = 90 |
| Software Name | Purpose |
|---|---|
| SBC-Collect | data collection |
| MOLREP | phasing |
| PHENIX | refinement |
| HKL-3000 | data reduction |
| HKL-3000 | data scaling |