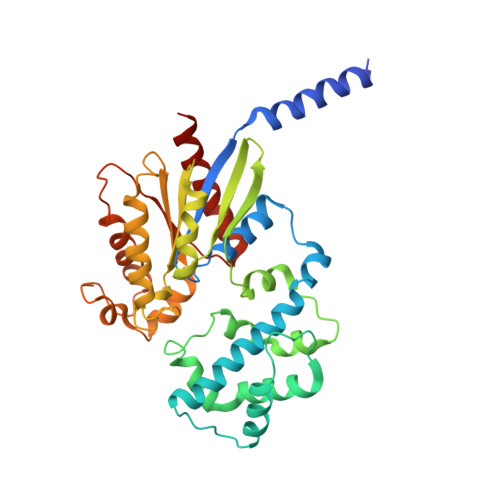
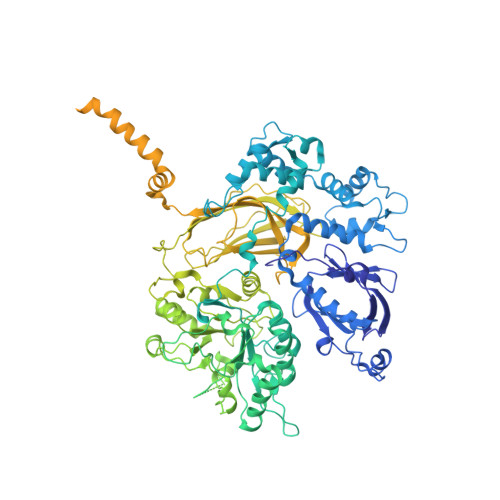
Full-length G alpha (q)-phospholipase C-beta 3 structure reveals interfaces of the C-terminal coiled-coil domain.
Lyon, A.M., Dutta, S., Boguth, C.A., Skiniotis, G., Tesmer, J.J.(2013) Nat Struct Mol Biol 20: 355-362
- PubMed: 23377541
- DOI: https://doi.org/10.1038/nsmb.2497
- Primary Citation of Related Structures:
4GNK - PubMed Abstract:
Phospholipase C-β (PLCβ) is directly activated by Gαq, but the molecular basis for how its distal C-terminal domain (CTD) contributes to maximal activity is poorly understood. Herein we present both the crystal structure and cryo-EM three-dimensional reconstructions of human full-length PLCβ3 in complex with mouse Gαq. The distal CTD forms an extended monomeric helical bundle consisting of three antiparallel segments with structural similarity to membrane-binding bin-amphiphysin-Rvs (BAR) domains. Sequence conservation of the distal CTD suggests putative membrane and protein interaction sites, the latter of which bind the N-terminal helix of Gαq in both the crystal structure and cryo-EM reconstructions. Functional analysis suggests that the distal CTD has roles in membrane targeting and in optimizing the orientation of the catalytic core at the membrane for maximal rates of lipid hydrolysis.
Organizational Affiliation:
Life Sciences Institute, University of Michigan, Ann Arbor, Michigan, USA.