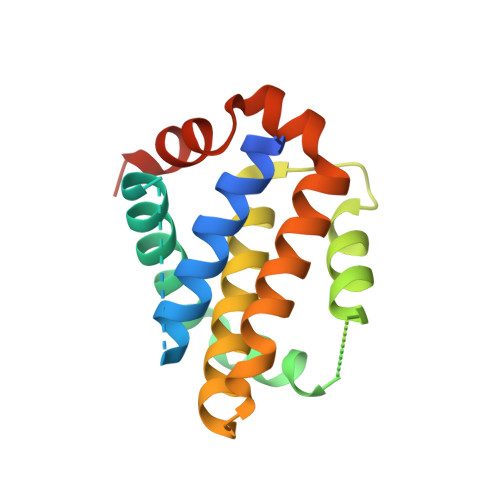
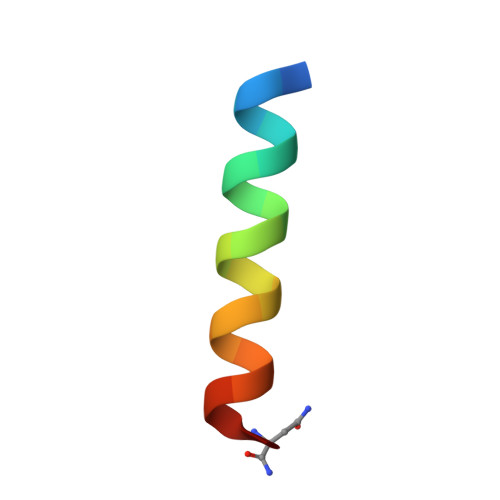
Rational design of proteolytically stable, cell-permeable peptide-based selective Mcl-1 inhibitors.
Muppidi, A., Doi, K., Edwardraja, S., Drake, E.J., Gulick, A.M., Wang, H.G., Lin, Q.(2012) J Am Chem Soc 134: 14734-14737
- PubMed: 22920569
- DOI: https://doi.org/10.1021/ja306864v
- Primary Citation of Related Structures:
4G35 - PubMed Abstract:
Direct chemical modifications provide a simple and effective means to "translate" bioactive helical peptides into potential therapeutics targeting intracellular protein-protein interactions. We previously showed that distance-matching bisaryl cross-linkers can reinforce peptide helices containing two cysteines at the i and i+7 positions and confer cell permeability to the cross-linked peptides. Here we report the first crystal structure of a biphenyl-cross-linked Noxa peptide in complex with its target Mcl-1 at 2.0 Å resolution. Guided by this structure, we remodeled the surface of this cross-linked peptide through side-chain substitution and N-methylation and obtained a pair of cross-linked peptides with substantially increased helicity, cell permeability, proteolytic stability, and cell-killing activity in Mcl-1-overexpressing U937 cells.
Organizational Affiliation:
Department of Chemistry, State University of New York at Buffalo, Buffalo, New York 14260-3000, USA.