Structure of Dengue Virus Ediii in Complex with Fab 2D73
Li, J., Watterson, D., Chang, C.W., Li, X.Q., Ericsson, D.J., Qiu, L.W., Cai, J.P., Chen, J., Fry, S.R., Cooper, M.A., Che, X.Y., Young, P.R., Kobe, B.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| ENVELOPE PROTEIN | 101 | dengue virus type 4 | Mutation(s): 0 | 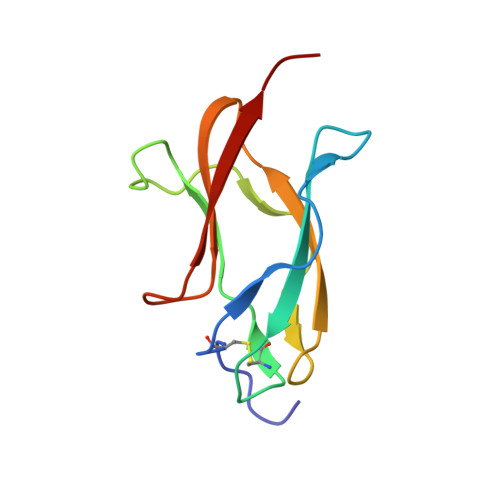 | |
UniProt | |||||
Find proteins for Q7TGC7 (dengue virus type 4) Explore Q7TGC7 Go to UniProtKB: Q7TGC7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q7TGC7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| FAB 2D73 HEAVY CHAIN | B [auth H] | 221 | Mus musculus | Mutation(s): 0 | 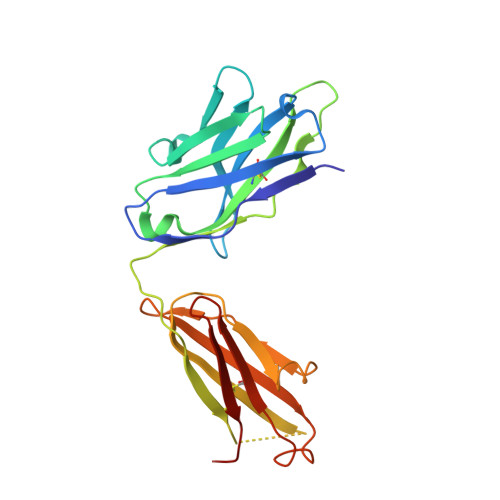 |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| FAB 2D73 LIGHT CHAIN | C [auth L] | 212 | Mus musculus | Mutation(s): 0 | 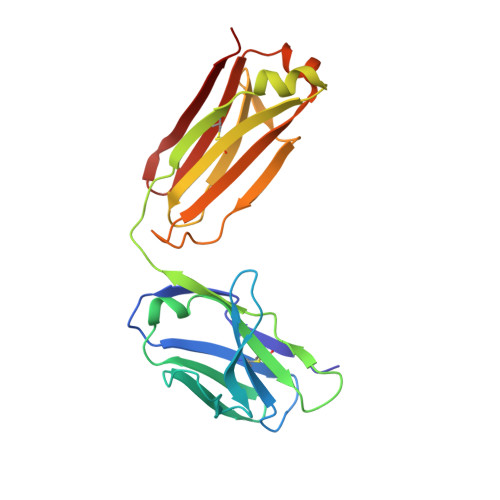 |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| PEG Query on PEG | K [auth H] | DI(HYDROXYETHYL)ETHER C4 H10 O3 MTHSVFCYNBDYFN-UHFFFAOYSA-N |  | ||
| GOL Query on GOL | L [auth H], M [auth H], R [auth L] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| CL Query on CL | H, N [auth L], O [auth L] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| NA Query on NA | D [auth A] E [auth A] F [auth A] G [auth H] I [auth H] | SODIUM ION Na FKNQFGJONOIPTF-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 108.655 | α = 90 |
| b = 108.655 | β = 90 |
| c = 108.564 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XDS | data reduction |
| SCALA | data scaling |
| PHASER | phasing |