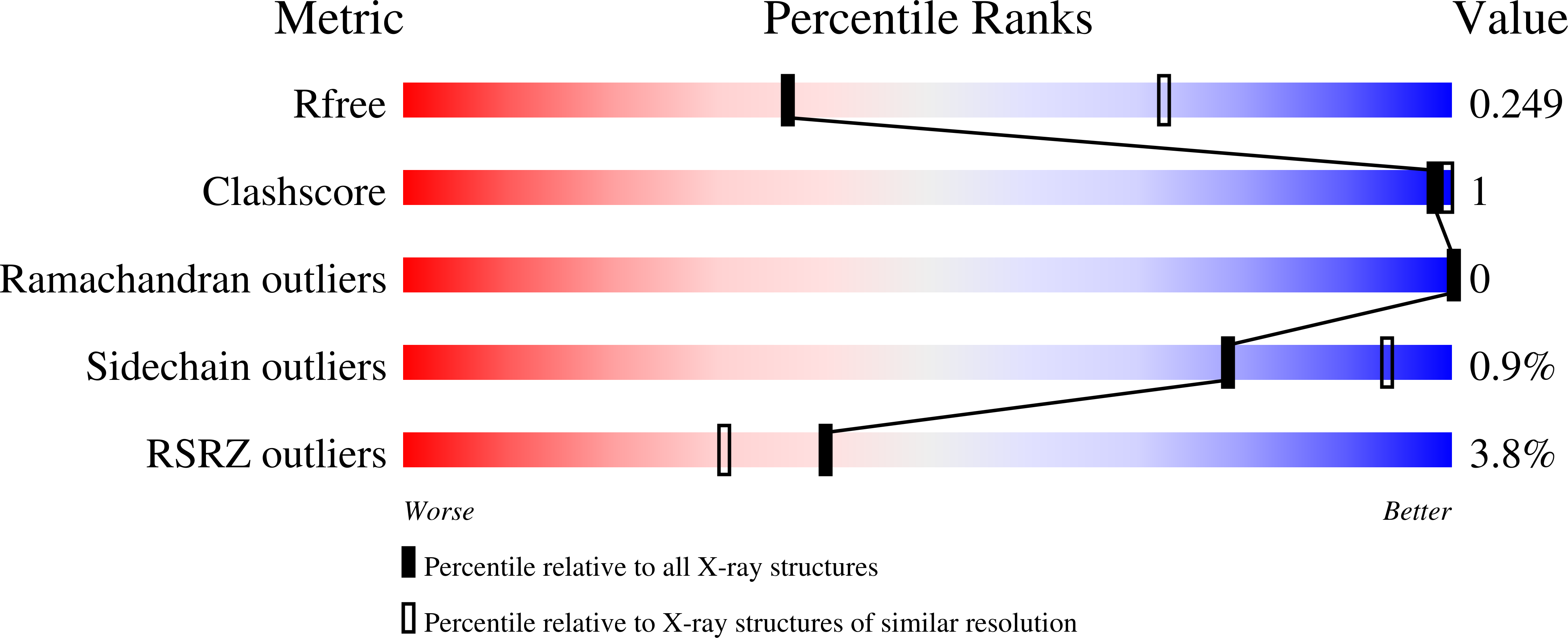
Enthalpy/Entropy Compensation Effects from Cavity Desolvation Underpin Broad Ligand Binding Selectivity for Rat Odorant Binding Protein 3
Portman, K.L., Long, J., Carr, S., Briand, L., Winzor, D.J., Searle, M.S., Scott, D.J.(2014) Biochemistry 53: 2371
- PubMed: 24665925
- DOI: https://doi.org/10.1021/bi5002344
- Primary Citation of Related Structures:
3ZQ3 - PubMed Abstract:
Evolution has produced proteins with exquisite ligand binding specificity, and manipulating this effect has been the basis for much of modern rational drug design. However, there are general classes of proteins with broader ligand selectivity linked to function, the origin of which is poorly understood. The odorant binding proteins (OBPs) sequester volatile molecules for transportation to the olfactory receptors. Rat OBP3, which we characterize by X-ray crystallography and NMR, binds a homologous series of aliphatic γ-lactones within its aromatic-rich hydrophobic pocket with remarkably little variation in affinity but extensive enthalpy/entropy compensation effects. We show that the binding energetics are modulated by two desolvation processes with quite different thermodynamic signatures. Ligand desolvation follows the classical hydrophobic effect; however, cavity desolvation is consistent with the liberation of "high energy" water molecules back into bulk solvent with a strong, but compensated, enthalpic contribution, which together underpin the origins of broad ligand binding selectivity.
Organizational Affiliation:
National Centre for Macromolecular Hydrodynamics, School of Biosciences, University of Nottingham , Sutton Bonington LE12 5RD, United Kingdom.