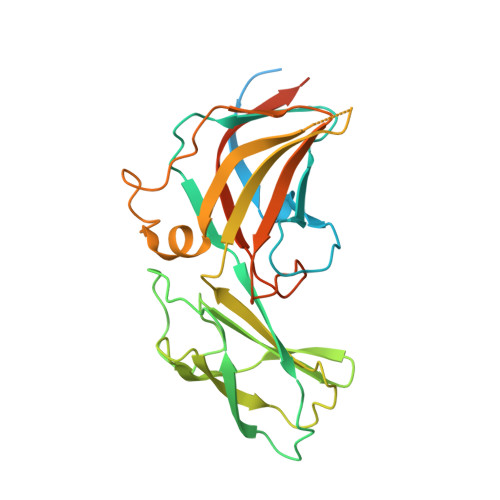
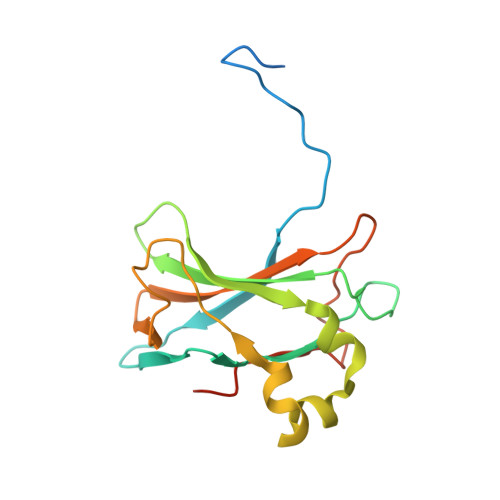
Bacteriophage P23-77 Capsid Protein Structures Reveal the Archetype of an Ancient Branch from a Major Virus Lineage.
Rissanen, I., Grimes, J.M., Pawlowski, A., Mantynen, S., Harlos, K., Bamford, J.K.H., Stuart, D.I.(2013) Structure 21: 718
- PubMed: 23623731
- DOI: https://doi.org/10.1016/j.str.2013.02.026
- Primary Citation of Related Structures:
3ZMN, 3ZMO, 3ZN4, 3ZN5, 3ZN6 - PubMed Abstract:
It has proved difficult to classify viruses unless they are closely related since their rapid evolution hinders detection of remote evolutionary relationships in their genetic sequences. However, structure varies more slowly than sequence, allowing deeper evolutionary relationships to be detected. Bacteriophage P23-77 is an example of a newly identified viral lineage, with members inhabiting extreme environments. We have solved multiple crystal structures of the major capsid proteins VP16 and VP17 of bacteriophage P23-77. They fit the 14 Å resolution cryo-electron microscopy reconstruction of the entire virus exquisitely well, allowing us to propose a model for both the capsid architecture and viral assembly, quite different from previously published models. The structures of the capsid proteins and their mode of association to form the viral capsid suggest that the P23-77-like and adeno-PRD1 lineages of viruses share an extremely ancient common ancestor.
Organizational Affiliation:
Department of Biological and Environmental Science and Nanoscience Center, University of Jyväskylä, Jyväskylä 40014, Finland.