An Unusual Arrangement of Two 14-3-3-Like Domains in the Smg5-Smg7 Heterodimer is Required for Efficient Nonsense-Mediated Mrna Decay.
Jonas, S., Weichenrieder, O., Izaurralde, E.(2013) Genes Dev 27: 211
- PubMed: 23348841
- DOI: https://doi.org/10.1101/gad.206672.112
- Primary Citation of Related Structures:
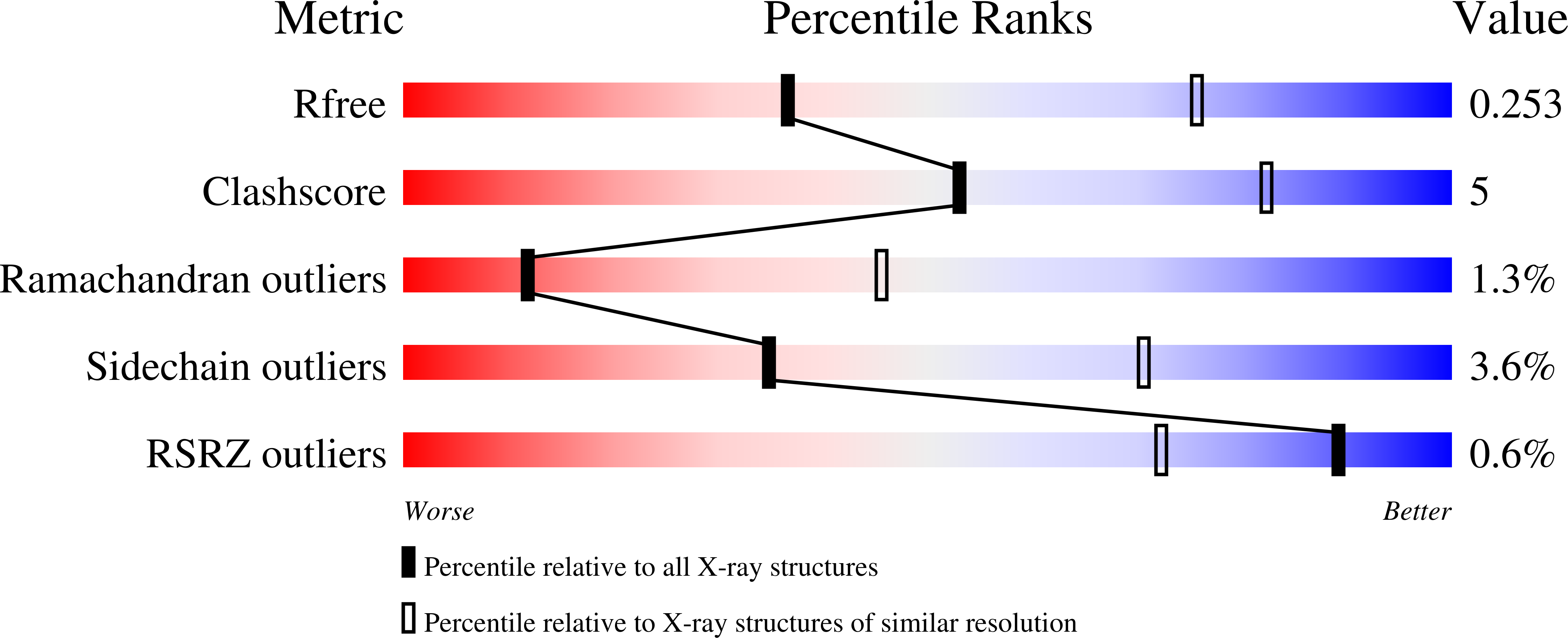
3ZHE - PubMed Abstract:
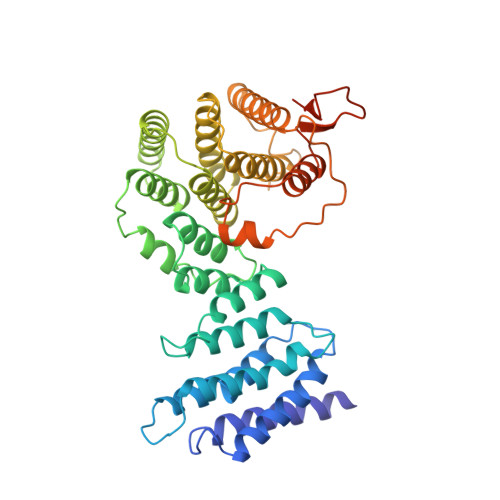
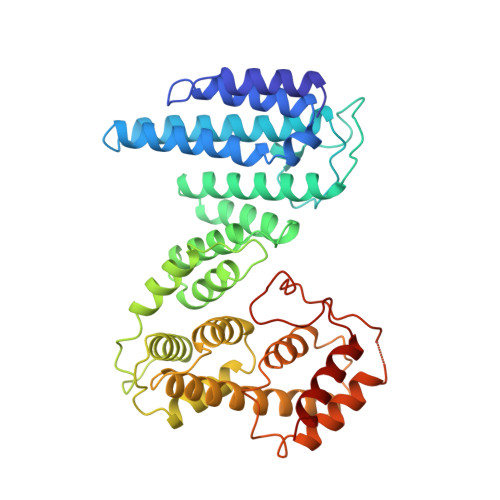
The nonsense-mediated mRNA decay (NMD) pathway triggers the rapid degradation of aberrant mRNAs containing premature translation termination codons (PTCs). In metazoans, NMD requires three 14-3-3-like proteins: SMG5, SMG6, and SMG7. These proteins are recruited to PTC-containing mRNAs through the interaction of their 14-3-3-like domains with phosphorylated UPF1, the central NMD effector. Recruitment of SMG5, SMG6, and SMG7 causes NMD target degradation. In this study, we report the crystal structure of the Caenorhabditis elegans SMG5-SMG7 complex. The 14-3-3-like phosphopeptide recognition domains of SMG5 and SMG7 heterodimerize in an unusual perpendicular back-to-back orientation in which the peptide-binding sites face opposite directions. Structure-based mutants and functional assays indicate that the SMG5-SMG7 interaction is conserved and is crucial for efficient NMD in human cells. Notably, we demonstrate that heterodimerization increases the affinity of the SMG5-SMG7 complex for UPF1. Furthermore, we show that the degradative activity of the SMG5-SMG7 complex resides in SMG7 and that the SMG5-SMG7 complex and SMG6 play partially redundant roles in the degradation of aberrant mRNAs. We propose that the SMG5-SMG7 complex binds to phosphorylated UPF1 with high affinity and recruits decay factors to the mRNA target through SMG7, thus promoting target degradation.
Organizational Affiliation:
Department of Biochemistry, Max Planck Institute for Developmental Biology, Tübingen, Germany.