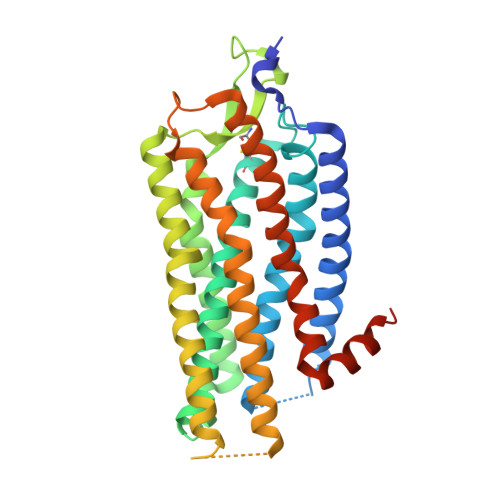
Structure of Signaling-Competent Neurotensin Receptor 1 Obtained by Directed Evolution in Escherichia Coli
Egloff, P., Hillenbrand, M., Klenk, C., Batyuk, A., Heine, P., Balada, S., Schlinkmann, K.M., Scott, D.J., Schuetz, M., Plueckthun, A.(2014) Proc Natl Acad Sci U S A 111: E655
- PubMed: 24453215
- DOI: https://doi.org/10.1073/pnas.1317903111
- Primary Citation of Related Structures:
3ZEV, 4BUO, 4BV0, 4BWB - PubMed Abstract:
Crystallography has advanced our understanding of G protein-coupled receptors, but low expression levels and instability in solution have limited structural insights to very few selected members of this large protein family. Using neurotensin receptor 1 (NTR1) as a proof of principle, we show that two directed evolution technologies that we recently developed have the potential to overcome these problems. We purified three neurotensin-bound NTR1 variants from Escherichia coli and determined their X-ray structures at up to 2.75 Å resolution using vapor diffusion crystallization experiments. A crystallized construct was pharmacologically characterized and exhibited ligand-dependent signaling, internalization, and wild-type-like agonist and antagonist affinities. Our structures are fully consistent with all biochemically defined ligand-contacting residues, and they represent an inactive NTR1 state at the cytosolic side. They exhibit significant differences to a previously determined NTR1 structure (Protein Data Bank ID code 4GRV) in the ligand-binding pocket and by the presence of the amphipathic helix 8. A comparison of helix 8 stability determinants between NTR1 and other crystallized G protein-coupled receptors suggests that the occupancy of the canonical position of the amphipathic helix is reduced to various extents in many receptors, and we have elucidated the sequence determinants for a stable helix 8. Our analysis also provides a structural rationale for the long-known effects of C-terminal palmitoylation reactions on G protein-coupled receptor signaling, receptor maturation, and desensitization.
Organizational Affiliation:
Department of Biochemistry, University of Zurich, 8057 Zurich, Switzerland.