Structural insight into equine lentivirus receptor 1
Qian, L., Han, X.D., Liu, X.Q.(2015) Protein Sci 24: 633-642
- PubMed: 25559821
- DOI: https://doi.org/10.1002/pro.2634
- Primary Citation of Related Structures:
3WVT - PubMed Abstract:
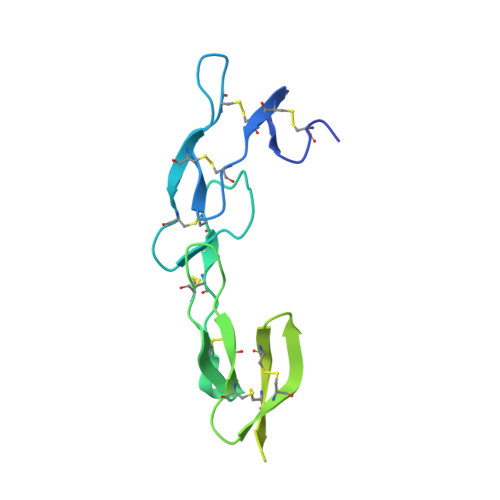
Equine lentivirus receptor 1 (ELR1) has been identified as a functional cellular receptor for equine infectious anemia virus (EIAV). Herein, recombinant ELR1 and EIAV surface glycoprotein gp90 were respectively expressed in Drosophila melanogaster S2 cells, and purified to homogeneity by Ni-NTA affinity chromatography and gel filtration chromatography. Gel filtration chromatography and analytical ultracentrifugation (AUC) analyses indicated that both ELR1 and gp90 existed as individual monomers in solution and formed a complex with a stoichiometry of 1:1 when mixed. The structure of ELR1 was first determined with the molecular replacement method, which belongs to the space group P42 21 2 with one molecule in an asymmetric unit. It contains eight antiparallel β-sheets, of which four are in cysteine rich domain 1 (CRD1) and two are in CRD2 and CRD3, respectively. Alignment of ELR1 with HVEM and CD134 indicated that Tyr61, Leu70, and Gly72 in CRD1 of ELR1 are important residues for binding to gp90. Isothermal titration calorimetry (ITC) experiments further confirmed that Leu70 and Gly72 are the critical residues.
Organizational Affiliation:
State Key Laboratory of Medicinal Chemical Biology, College of Life Sciences, Nankai University, Tianjin, 300071, China.