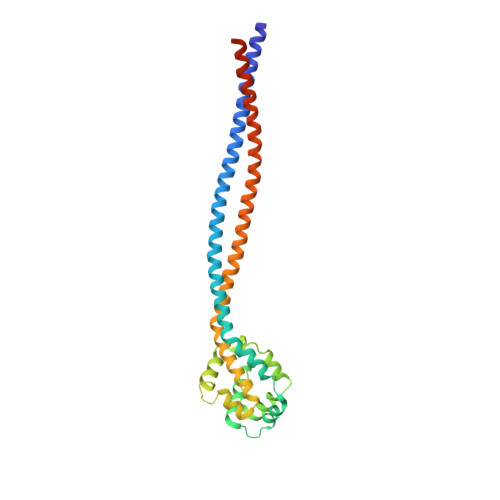
Structure of the entire stalk region of the Dynein motor domain
Nishikawa, Y., Oyama, T., Kamiya, N., Kon, T., Toyoshima, Y.Y., Nakamura, H., Kurisu, G.(2014) J Mol Biol 426: 3232-3245
- PubMed: 25058684
- DOI: https://doi.org/10.1016/j.jmb.2014.06.023
- Primary Citation of Related Structures:
3WUQ - PubMed Abstract:
Dyneins are large microtubule-based motor complexes that power a range of cellular processes including the transport of organelles, as well as the beating of cilia and flagella. The motor domain is located within the dynein heavy chain and comprises an N-terminal mechanical linker element, a central ring of six AAA+ modules of which four bind or hydrolyze ATP, and a long stalk extending from the AAA+ring with a microtubule-binding domain (MTBD) at its tip. A crucial mechanism underlying the motile activity of cytoskeletal motor proteins is precise coupling between the ATPase and track-binding activities. In dynein, a stalk region consisting of a long (~15nm) antiparallel coiled coil separates these two activities, which must facilitate communication between them. This communication is mediated by a small degree of helix sliding in the coiled coil. However, no high-resolution structure is available of the entire stalk region including the MTBD. Here, we have reported the structure of the entire stalk region of mouse cytoplasmic dynein in a weak microtubule-binding state, which was determined using X-ray crystallography, and have compared it with the dynein motor domain from Dictyostelium discoideum in a strong microtubule-binding state and with a mouse MTBD with its distal portion of the coiled coil fused to seryl-tRNA synthetase from Thermus thermophilus. Our results strongly support the helix-sliding model based on the complete structure of the dynein stalk with a different form of coiled-coil packing. We also propose a plausible mechanism of helix sliding together with further analysis using molecular dynamics simulations. Our results present the importance of conserved proline residues for an elastic motion of stalk coiled coil and imply the manner of change between high-affinity state and low-affinity state of MTBD.
Organizational Affiliation:
Institute for Protein Research, Osaka University, 3-2 Yamadaoka, Suita, Osaka 565-0871, Japan; Department of Macromolecular Science, Graduate School of Science, Osaka University, Toyonaka, Osaka 560-0043, Japan.