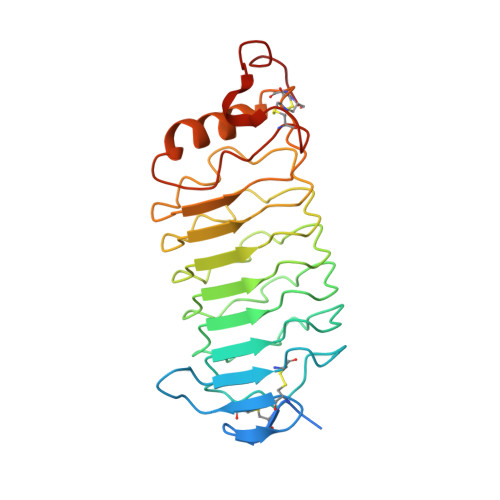
Crystal structure of the lamprey variable lymphocyte receptor C reveals an unusual feature in its N-terminal capping module.
Kanda, R., Sutoh, Y., Kasamatsu, J., Maenaka, K., Kasahara, M., Ose, T.(2014) PLoS One 9: e85875-e85875
- PubMed: 24465760
- DOI: https://doi.org/10.1371/journal.pone.0085875
- Primary Citation of Related Structures:
3WO9 - PubMed Abstract:
Jawless vertebrates represented by lampreys and hagfish use variable lymphocyte receptors (VLRs) as antigen receptors to mount adaptive immune responses. VLRs generate diversity that is comparable to immunoglobulins and T-cell receptors by a gene conversion-like mechanism, which is mediated by cytosine deaminases. Currently, three types of VLRs, VLRA, VLRB, and VLRC, have been identified in lampreys. Crystal structures of VLRA and VLRB in complex with antigens have been reported recently, but no structural information is available for VLRC. Here, we present the first crystal structure of VLRC from the Japanese lamprey (Lethenteron japonicum). Similar to VLRA and VLRB, VLRC forms a typical horseshoe-like solenoid structure with a variable concave surface. Strikingly, its N-terminal cap has a long loop with limited sequence variability that protrudes toward the concave surface, which is the putative antigen-binding surface. Furthermore, as predicted previously, its C-terminal cap lacks a highly variable protruding loop that plays an important role in antigen recognition by lamprey VLRA and VLRB. Recent work suggests that VLRC+ lymphocytes in jawless vertebrates might be akin to γδ T cells in jawed vertebrates. Structural features of lamprey VLRC described here suggest that it may recognize antigens in a unique manner.
Organizational Affiliation:
Graduate School of Life Sciences, Hokkaido University, Sapporo, Japan.