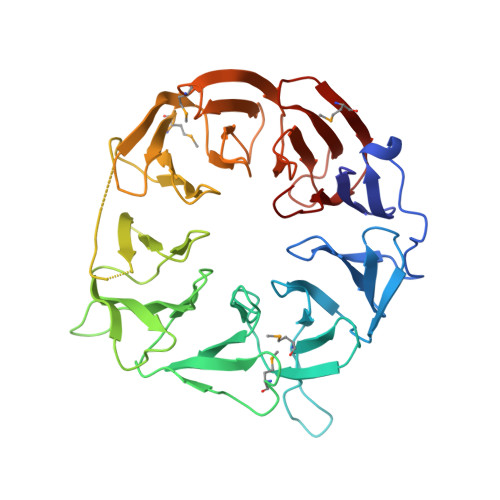
Crystal structure of the eukaryotic translation initiation factor 2A from Schizosaccharomyces pombe.
Kashiwagi, K., Ito, T., Yokoyama, S.(2014) J Struct Funct Genomics 15: 125-130
- PubMed: 24569939
- DOI: https://doi.org/10.1007/s10969-014-9177-y
- Primary Citation of Related Structures:
3WJ9 - PubMed Abstract:
The eukaryotic translation initiation factor 2A (eIF2A) was identified as a factor that stimulates the binding of methionylated initiator tRNA (Met-tRNA i (Met) ) to the 40S ribosomal subunit, but its physiological role remains poorly defined. Recently, eIF2A was shown to be involved in unconventional translation initiation from CUG codons and in viral protein synthesis under stress conditions where eIF2 is inactivated. We determined the crystal structure of the WD-repeat domain of Schizosaccharomyces pombe eIF2A at 2.5 Å resolution. The structure adopts a novel nine-bladed β-propeller fold. In contrast to the usual β-propeller proteins, the central channel of the molecule has the narrower opening on the bottom of the protein and the wider opening on the top. Highly conserved residues are concentrated in the positively-charged top face, suggesting the importance of this face for interactions with nucleic acids or other initiation factors.
Organizational Affiliation:
Department of Biophysics and Biochemistry, Graduate School of Science, The University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo, 113-0033, Japan.