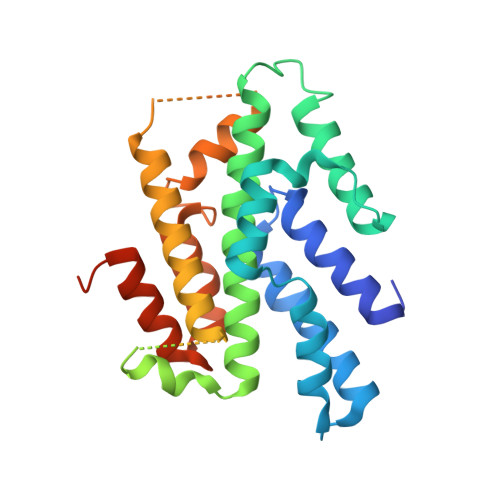
Crystal structure of the homology domain of the eukaryotic DNA replication proteins sld3/treslin.
Itou, H., Muramatsu, S., Shirakihara, Y., Araki, H.(2014) Structure 22: 1341-1347
- PubMed: 25126958
- DOI: https://doi.org/10.1016/j.str.2014.07.001
- Primary Citation of Related Structures:
3WI3 - PubMed Abstract:
The initiation of eukaryotic chromosomal DNA replication requires the formation of an active replicative helicase at the replication origins of chromosomal DNA. Yeast Sld3 and its metazoan counterpart Treslin are the hub proteins mediating protein associations critical for the helicase formation. Here, we show the crystal structure of the central domain of Sld3 that is conserved in Sld3/Treslin family of proteins. The domain consists of two segments with 12 helices and is sufficient to bind to Cdc45, the essential helicase component. The structure model of the Sld3-Cdc45 complex, which is crucial for the formation of the active helicase, is proposed.
Organizational Affiliation:
Structural Biology Center, National Institute of Genetics, Yata1111, Mishima, Shizuoka 411-8540, Japan; Department of Genetics, SOKENDAI, Yata1111, Mishima, Shizuoka 411-8540, Japan. Electronic address: hitou@nig.ac.jp.