Structure of the quinoline N-hydroxylating cytochrome P450 RauA, an essential enzyme that confers antibiotic activity on aurachin alkaloids
Yasutake, Y., Kitagawa, W., Hata, M., Nishioka, T., Ozaki, T., Nishiyama, M., Kuzuyama, T., Tamura, T.(2014) FEBS Lett 588: 105-110
- PubMed: 24269679
- DOI: https://doi.org/10.1016/j.febslet.2013.11.016
- Primary Citation of Related Structures:
3WEC - PubMed Abstract:
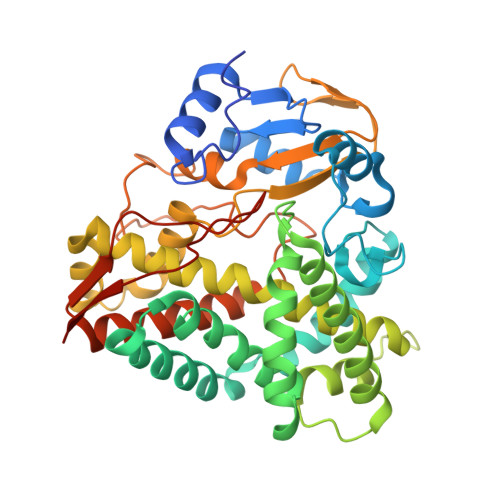
The cytochrome P450 RauA from Rhodococcus erythropolis JCM 6824 catalyzes the hydroxylation of a nitrogen atom in the quinolone ring of aurachin, thereby conferring strong antibiotic activity on the aurachin alkaloid. Here, we report the crystal structure of RauA in complex with its substrate, a biosynthetic intermediate of aurachin RE. Clear electron density showed that the quinolone ring is oriented parallel to the porphyrin plane of the heme cofactor, while the farnesyl chain curls into a U-shape topology and is buried inside the solvent-inaccessible hydrophobic interior of RauA. The nearest atom from the heme iron is the quinolone nitrogen (4.3Å), which is consistent with RauA catalyzing the N-hydroxylation of the quinolone ring to produce mature aurachin RE.
Organizational Affiliation:
Bioproduction Research Institute, National Institute of Advanced Industrial Science and Technology (AIST), 2-17-2-1 Tsukisamu-Higashi, Toyohira-ku, Sapporo 062-8517, Japan. Electronic address: y-yasutake@aist.go.jp.