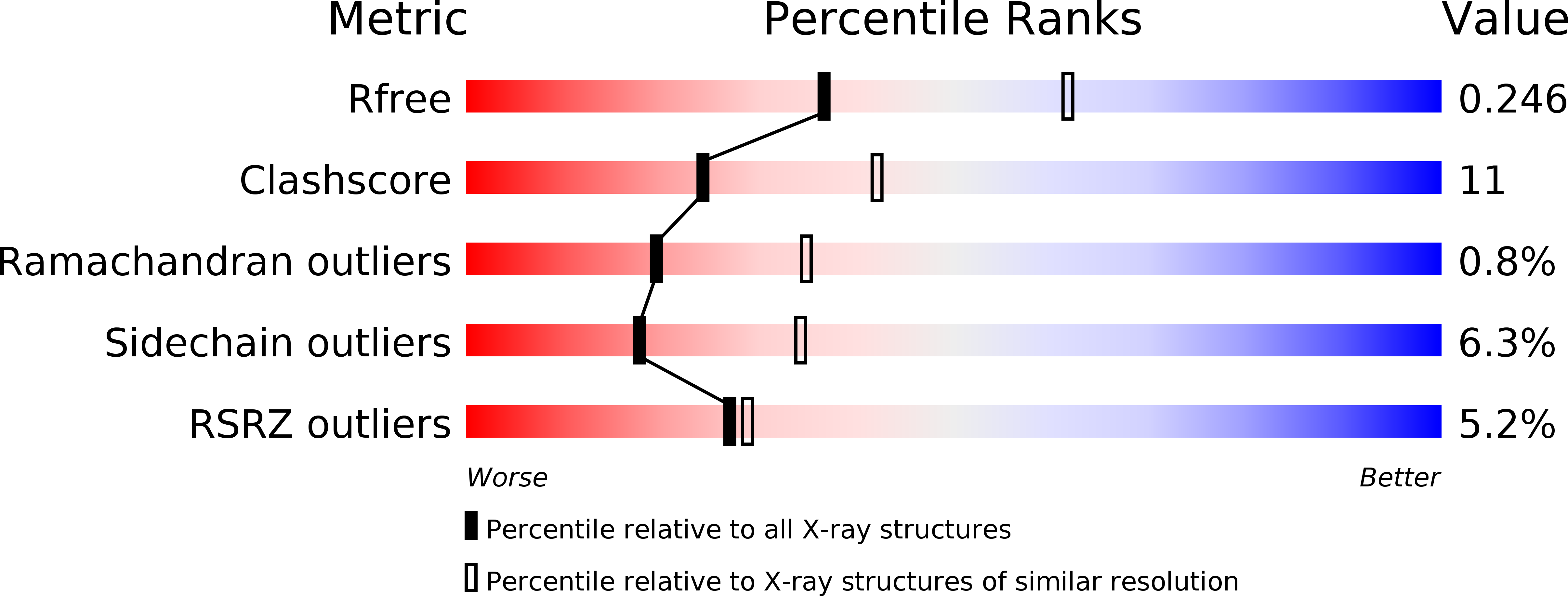
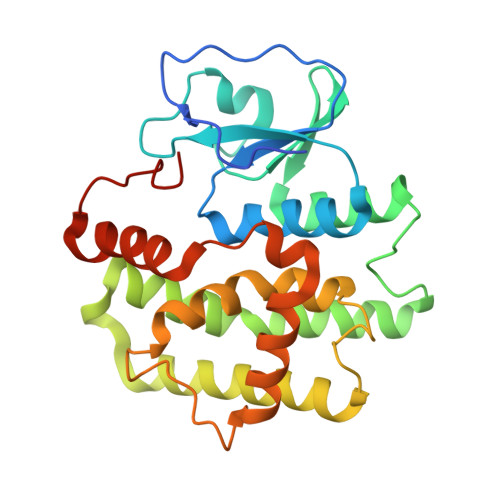
Three-dimensional structure of a Bombyx mori Omega-class glutathione transferase.
Yamamoto, K., Suzuki, M., Higashiura, A., Nakagawa, A.(2013) Biochem Biophys Res Commun 438: 588-593
- PubMed: 23939046
- DOI: https://doi.org/10.1016/j.bbrc.2013.08.011
- Primary Citation of Related Structures:
3WD6 - PubMed Abstract:
Glutathione transferases (GSTs) are major phase II detoxification enzymes that play central roles in the defense against various environmental toxicants as well as oxidative stress. Here we report the crystal structure of an Omega-class glutathione transferase of Bombyx mori, bmGSTO, to gain insight into its catalytic mechanism. The structure of bmGSTO complexed with glutathione determined at a resolution of 2.5Å reveals that it exists as a dimer and is structurally similar to Omega-class GSTs with respect to its secondary and tertiary structures. Analysis of a complex between bmGSTO and glutathione showed that bound glutathione was localized to the glutathione-binding site (G-site). Site-directed mutagenesis of bmGSTO mutants indicated that amino acid residues Leu62, Lys65, Lys77, Val78, Glu91 and Ser92 in the G-site contribute to catalytic activity.
Organizational Affiliation:
Faculty of Agriculture, Kyushu University Graduate School, 6-10-1 Hakozaki, Higashi-ku, Fukuoka 812-8581, Japan. yamamok@agr.kyushu-u.ac.jp