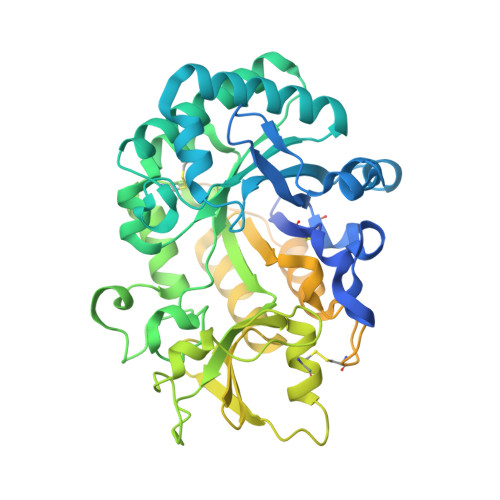
Structural characteristics of an insect group I chitinase, an enzyme indispensable to moulting
Chen, L., Liu, T., Zhou, Y., Chen, Q., Shen, X., Yang, Q.(2014) Acta Crystallogr D Biol Crystallogr 70: 932-942
- PubMed: 24699639
- DOI: https://doi.org/10.1107/S1399004713033841
- Primary Citation of Related Structures:
3W4R, 3WKZ, 3WL0, 3WL1 - PubMed Abstract:
Insects possess a greater number of chitinases than any other organisms. This work is the first report of unliganded and oligosaccharide-complexed crystal structures of the insect chitinase OfChtI from Ostrinia furnacalis, which is essential to moulting. The obtained crystal structures were solved at resolutions between 1.7 and 2.2 Å. A structural comparison with other chitinases revealed that OfChtI contains a long substrate-binding cleft similar to the bacterial chitinase SmChiB from Serratia marcescens. However, unlike the exo-acting SmChiB, which has a blocked and tunnel-like cleft, OfChtI possesses an open and groove-like cleft. The complexed structure of the catalytic domain of OfChtI (OfChtI-CAD) with (GlcNAc)2/3 indicates that the reducing sugar at subsite -1 is in an energetically unfavoured `boat' conformation, a state that possibly exists just before the completion of catalysis. Because OfChtI is known to act from nonreducing ends, (GlcNAc)3 would be a hydrolysis product of (GlcNAc)6, suggesting that OfChtI possesses an endo enzymatic activity. Furthermore, a hydrophobic plane composed of four surface-exposed aromatic residues is adjacent to the entrance to the substrate-binding cleft. Mutations of these residues greatly impair the chitin-binding activity, indicating that this hydrophobic plane endows OfChtI-CAD with the ability to anchor chitin. This work reveals the unique structural characteristics of an insect chitinase.
Organizational Affiliation:
School of Life Science and Biotechnology, Dalian University of Technology, 2 Linggong Road, Dalian, Liaoning 116024, People's Republic of China.