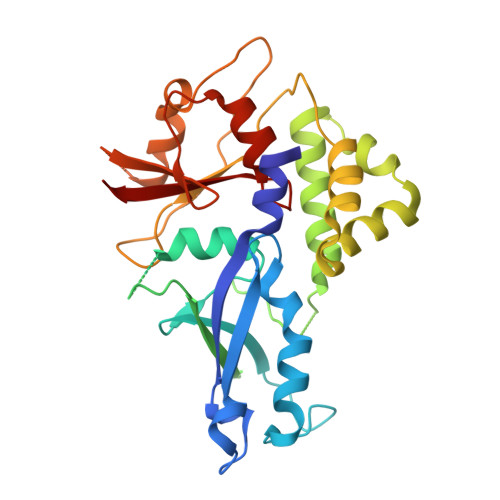
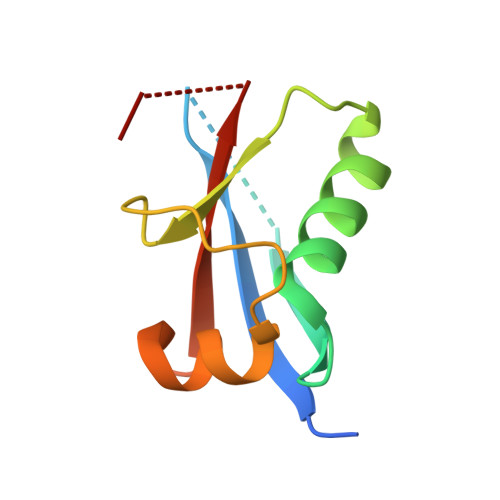
Structure of the Atg12-Atg5 conjugate reveals a platform for stimulating Atg8-PE conjugation
Noda, N.N., Fujioka, Y., Hanada, T., Ohsumi, Y., Inagaki, F.(2013) EMBO Rep 14: 206-211
- PubMed: 23238393
- DOI: https://doi.org/10.1038/embor.2012.208
- Primary Citation of Related Structures:
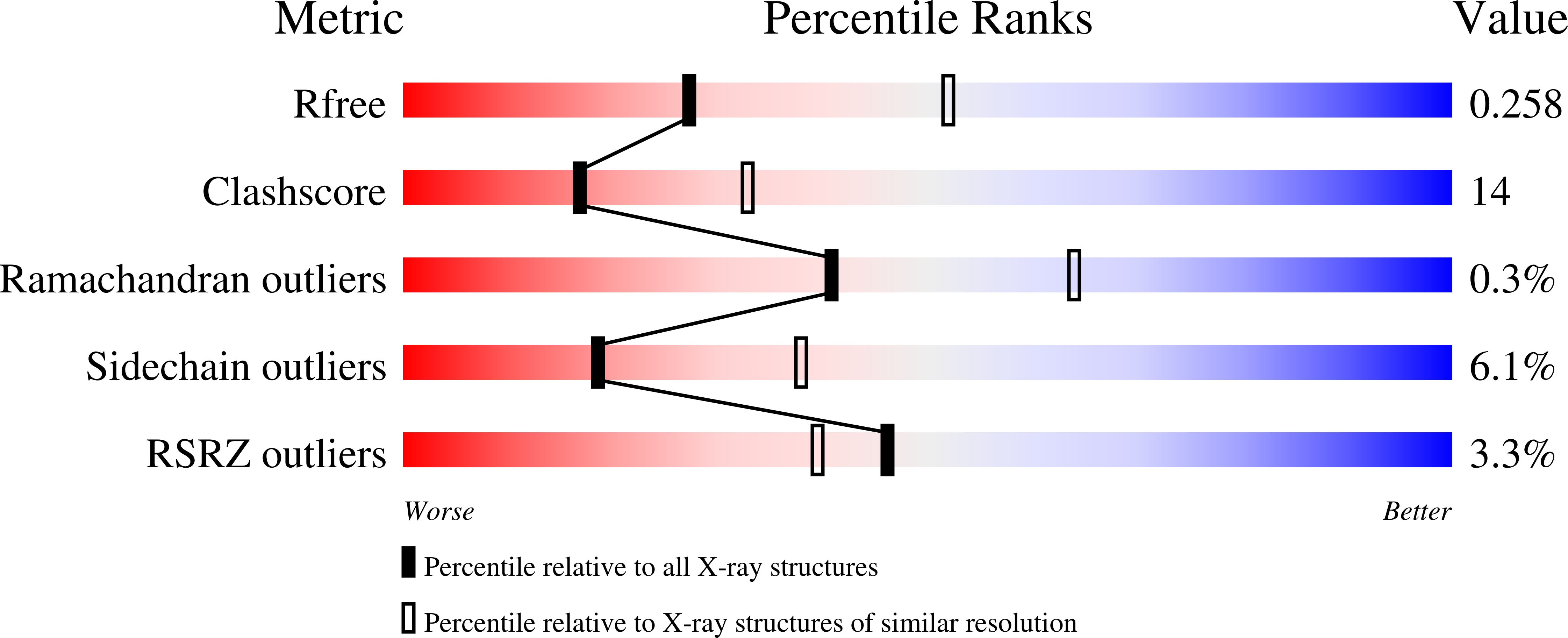
3W1S - PubMed Abstract:
Atg12 is conjugated to Atg5 through enzymatic reactions similar to ubiquitination. The Atg12-Atg5 conjugate functions as an E3-like enzyme to promote lipidation of Atg8, whereas lipidated Atg8 has essential roles in both autophagosome formation and selective cargo recognition during autophagy. However, the molecular role of Atg12 modification in these processes has remained elusive. Here, we report the crystal structure of the Atg12-Atg5 conjugate. In addition to the isopeptide linkage, Atg12 forms hydrophobic and hydrophilic interactions with Atg5, thereby fixing its position on Atg5. Structural comparison with unmodified Atg5 and mutational analyses showed that Atg12 modification neither induces a conformational change in Atg5 nor creates a functionally important architecture. Rather, Atg12 functions as a binding module for Atg3, the E2 enzyme for Atg8, thus endowing Atg5 with the ability to interact with Atg3 to facilitate Atg8 lipidation.
Organizational Affiliation:
Microbial Chemistry Research Foundation, Laboratory of Molecular Structure, Institute of Microbial Chemistry, 3-14-23, Kamiosaki, Shinagawa-ku, Tokyo 141-0021, Japan. nn@bikaken.or.jp