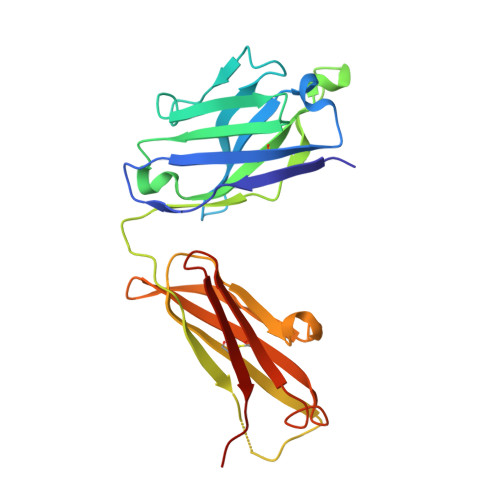
Antigenic switching of hepatitis B virus by alternative dimerization of the capsid protein.
Dimattia, M.A., Watts, N.R., Stahl, S.J., Grimes, J.M., Steven, A.C., Stuart, D.I., Wingfield, P.T.(2013) Structure 21: 133-142
- PubMed: 23219881
- DOI: https://doi.org/10.1016/j.str.2012.10.017
- Primary Citation of Related Structures:
3V6F, 3V6Z - PubMed Abstract:
Chronic hepatitis B virus (HBV) infection afflicts millions worldwide with cirrhosis and liver cancer. HBV e-antigen (HBeAg), a clinical marker for disease severity, is a nonparticulate variant of the protein (core antigen, HBcAg) that forms the building-blocks of capsids. HBeAg is not required for virion production, but is implicated in establishing immune tolerance and chronic infection. Here, we report the crystal structure of HBeAg, which clarifies how the short N-terminal propeptide of HBeAg induces a radically altered mode of dimerization relative to HBcAg (∼140° rotation), locked into place through formation of intramolecular disulfide bridges. This structural switch precludes capsid assembly and engenders a distinct antigenic repertoire, explaining why the two antigens are cross-reactive at the T cell level (through sequence identity) but not at the B cell level (through conformation). The structure offers insight into how HBeAg may establish immune tolerance for HBcAg while evading its robust immunogenicity.
Organizational Affiliation:
Division of Structural Biology, Henry Wellcome Building for Genomic Medicine, University of Oxford, Roosevelt Drive, Headington OX3 7BN, UK; Laboratory of Structural Biology Research, National Institute of Arthritis and Musculoskeletal and Skin Diseases; National Institutes of Health, Bethesda, MD 20892, USA.