Cyanobacterial Peptides as a Prototype for the Design of Potent beta-Secretase Inhibitors and the Development of Selective Chemical Probes for Other Aspartic Proteases
Liu, Y., Zhang, W., Li, L., Salvador, L.A., Chen, T.T., Chen, W.Y., Felsenstein, K.M., Ladd, T.B., Price, A.R., Golde, T.E., He, J., Xu, Y.C., Li, Y., Luesch, H.(2012) J Med Chem 55: 10749-10765
- PubMed: 23181502
- DOI: https://doi.org/10.1021/jm301630s
- Primary Citation of Related Structures:
3UQP, 3UQR, 4DV9, 4DVF, 4FGX - PubMed Abstract:
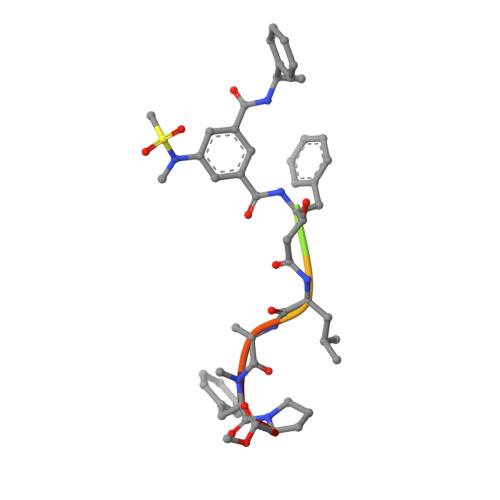
Inspired by marine cyanobacterial natural products, we synthesized modified peptides with a central statine-core unit, characteristic for aspartic protease inhibition. A series of tasiamide B analogues inhibited BACE1, a therapeutic target in Alzheimer's disease. We probed the stereospecificity of target engagement and determined additional structure-activity relationships with respect to BACE1 and related aspartic proteases, cathepsins D and E. We cocrystallized selected inhibitors with BACE1 to reveal the structural basis for the activity. Hybrid molecules that combine features of tasiamide B and an isophthalic acid moiety-containing sulfonamide showed nanomolar cellular activity. Compounds were screened in a series of rigorous complementary cell-based assays. We measured secreted APP ectodomain (sAPPβ), membrane bound carboxyl terminal fragment (CTF), levels of β-amyloid (Aβ) peptides and selectivity for β-secretase (BACE1) over γ-secretase. Prioritized compounds showed reasonable stability in vitro and in vivo, and our most potent inhibitor showed efficacy in reducing Aβ levels in the rodent brain.
Organizational Affiliation:
Department of Medicinal Chemistry, University of Florida, Gainesville, Florida 32610, United States.