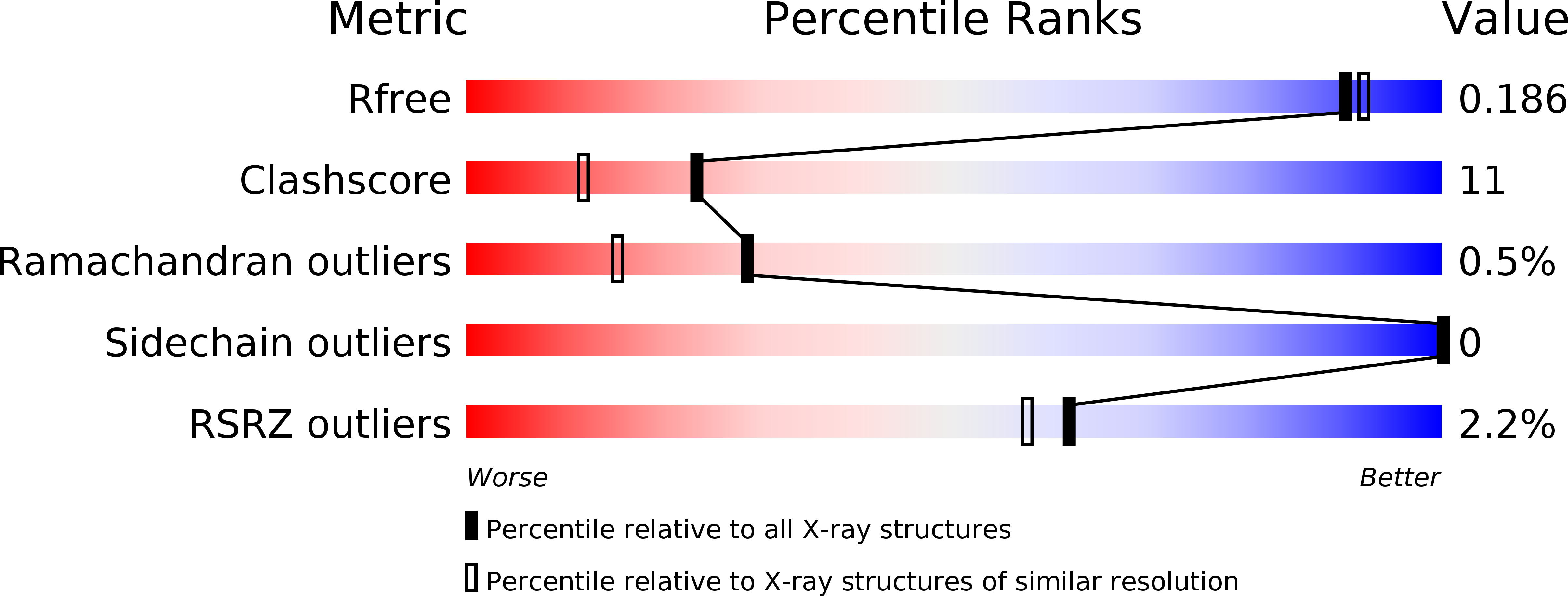
Covalent attachment of pyridoxal-phosphate derivatives to 14-3-3 proteins.
Roglin, L., Thiel, P., Kohlbacher, O., Ottmann, C.(2012) Proc Natl Acad Sci U S A 109: E1051-E1053
- PubMed: 22532669
- DOI: https://doi.org/10.1073/pnas.1116592109
- Primary Citation of Related Structures:
3U9X