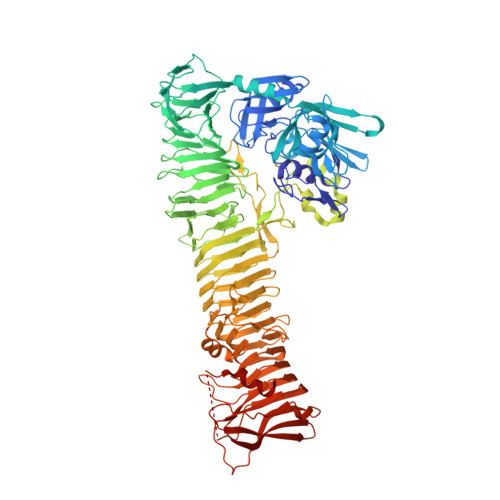
Crystal Structure of the Passenger Domain of the Escherichia coli Autotransporter EspP.
Khan, S., Mian, H.S., Sandercock, L.E., Chirgadze, N.Y., Pai, E.F.(2011) J Mol Biol 413: 985-1000
- PubMed: 21964244
- DOI: https://doi.org/10.1016/j.jmb.2011.09.028
- Primary Citation of Related Structures:
3SZE - PubMed Abstract:
Autotransporters represent a large superfamily of known and putative virulence factors produced by Gram-negative bacteria. They consist of an N-terminal "passenger domain" responsible for the specific effector functions of the molecule and a C-terminal "β-domain" responsible for translocation of the passenger across the bacterial outer membrane. Here, we present the 2.5-Å crystal structure of the passenger domain of the extracellular serine protease EspP, produced by the pathogen Escherichia coli O157:H7 and a member of the serine protease autotransporters of Enterobacteriaceae (SPATEs). Like the previously structurally characterized SPATE passenger domains, the EspP passenger domain contains an extended right-handed parallel β-helix preceded by an N-terminal globular domain housing the catalytic function of the protease. Of note, however, is the absence of a second globular domain protruding from this β-helix. We describe the structure of the EspP passenger domain in the context of previous results and provide an alternative hypothesis for the function of the β-helix within SPATEs.
Organizational Affiliation:
Campbell Family Cancer Research Institute, Ontario Cancer Institute, University Health Network, Toronto Medical Discovery Tower, Toronto, Ontario, Canada M5G 1L7.