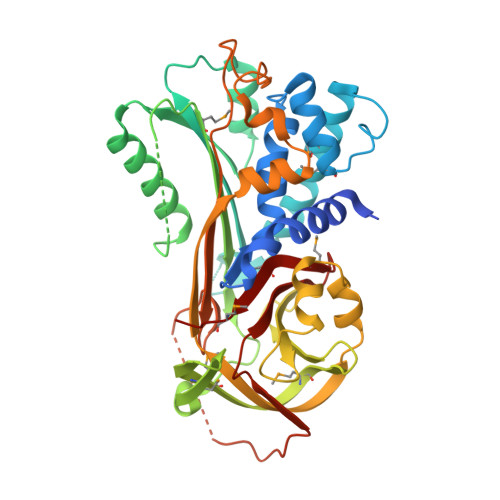
Three-dimensional structure of a schistosome serpin revealing an unusual configuration of the helical subdomain.
Granzin, J., Huang, Y., Topbas, C., Huang, W., Wu, Z., Misra, S., Hazen, S.L., Blanton, R.E., Lee, X., Weiergraber, O.H.(2012) Acta Crystallogr D Biol Crystallogr 68: 686-694
- PubMed: 22683791
- DOI: https://doi.org/10.1107/S0907444912008372
- Primary Citation of Related Structures:
3STO - PubMed Abstract:
Parasitic organisms are constantly challenged by the defence mechanisms of their respective hosts, which often depend on serine protease activities. Consequently, protease inhibitors such as those belonging to the serpin superfamily have emerged as protective elements that support the survival of the parasites. This report describes the crystal structure of ShSPI, a serpin from the trematode Schistosoma haematobium. The protein is exposed on the surface of invading cercaria as well as of adult worms, suggesting its involvement in the parasite-host interaction. While generally conforming to the well established serpin fold, the structure reveals several distinctive features, mostly concerning the helical subdomain of the protein. It is proposed that these peculiarities are related to the unique biological properties of a small serpin subfamily which is conserved among pathogenic schistosomes.
Organizational Affiliation:
Institute of Complex Systems, ICS-6: Structural Biochemistry, Forschungszentrum Jülich, 52425 Jülich, Germany.