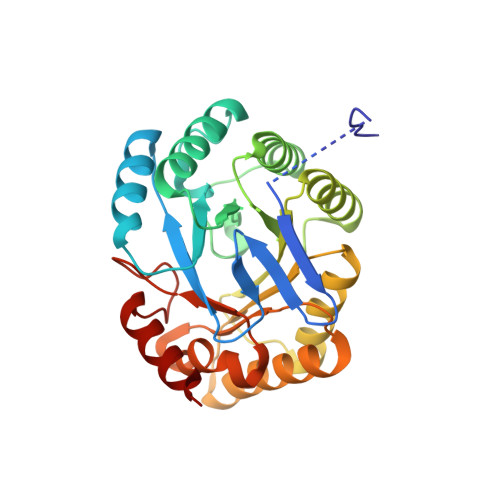
Crystal Structure of the 3-Dehydroquinate Dehydratase (aroD) from Salmonella enterica Typhimurium LT2 with Malonate and Boric Acid at the Active Site
Light, S.H., Minasov, G., Duban, M.-E., Halavaty, A.S., Krishna, S.N., Shuvalova, L., Kwon, K., Anderson, W.F., Center for Structural Genomics of Infectious Diseases (CSGID)To be published.