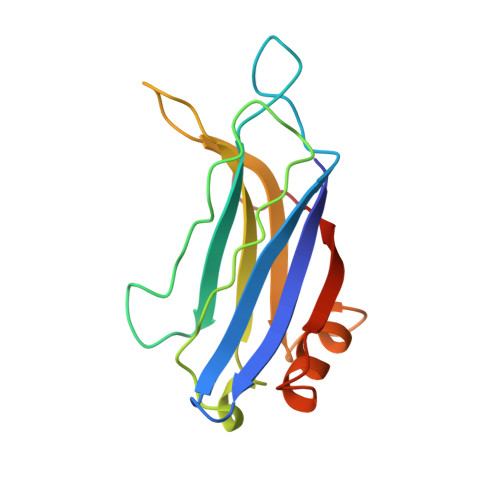
Structure of the Janus-faced C2B domain of rabphilin.
Ubach, J., Garcia, J., Nittler, M.P., Sudhof, T.C., Rizo, J.(1999) Nat Cell Biol 1: 106-112
- PubMed: 10559882
- DOI: https://doi.org/10.1038/10076
- Primary Citation of Related Structures:
3RPB - PubMed Abstract:
C2 domains are widespread protein modules that often occur as tandem repeats in many membrane-trafficking proteins such as synaptotagmin and rabphilin. The first and second C2 domains (C2A and C2B, respectively) have a high degree of homology but also specific differences. The structure of the C2A domain of synaptotagmin I has been extensively studied but little is known about the C2B domains. We have used NMR spectroscopy to determine the solution structure of the C2B domain of rabphilin. The overall structure of the C2B domain is very similar to that of other C2 domains, with a rigid beta-sandwich core and loops at the top (where Ca2+ binds) and the bottom. Surprisingly, a relatively long alpha-helix is inserted at the bottom of the domain and is conserved in all C2B domains. Our results, together with the Ca(2+)-independent interactions observed for C2B domains, indicate that these domains have a Janus-faced nature, with a Ca(2+)-binding top surface and a Ca(2+)-independent bottom surface.
Organizational Affiliation:
Department of Biochemistry, University of Texas Southwestern Medical Center, Dallas 75235, USA.