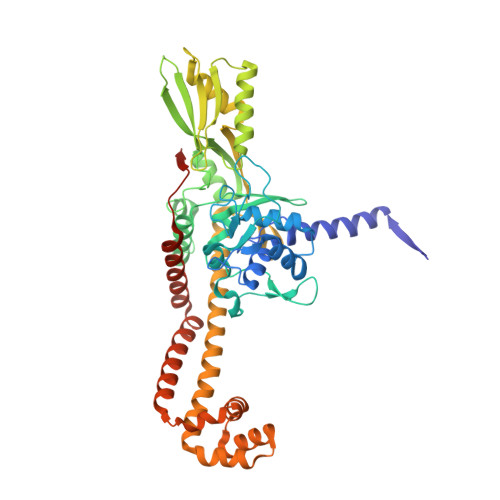
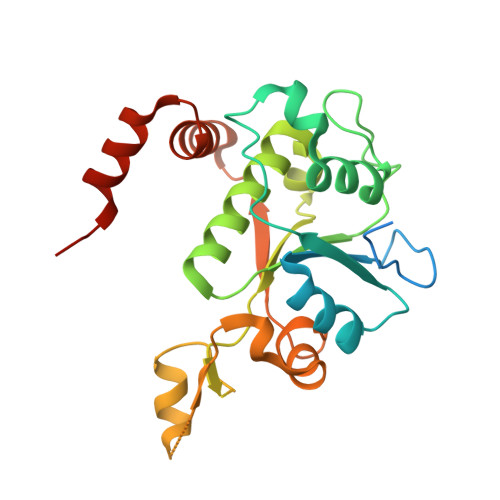
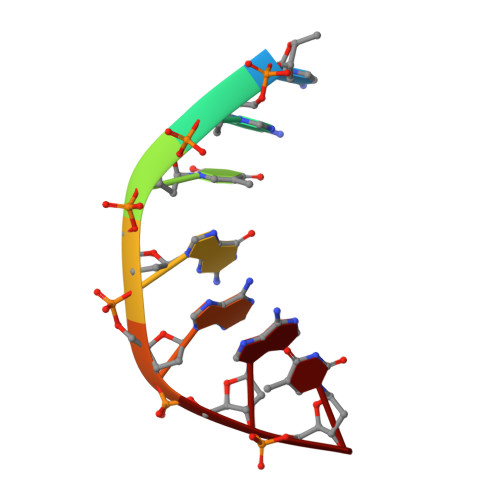
Structure of a quinolone-stabilized cleavage complex of topoisomerase IV from Klebsiella pneumoniae and comparison with a related Streptococcus pneumoniae complex.
Veselkov, D.A., Laponogov, I., Pan, X.S., Selvarajah, J., Skamrova, G.B., Branstrom, A., Narasimhan, J., Prasad, J.V., Fisher, L.M., Sanderson, M.R.(2016) Acta Crystallogr D Biol Crystallogr 72: 488-496
- PubMed: 27050128
- DOI: https://doi.org/10.1107/S2059798316001212
- Primary Citation of Related Structures:
3RAE, 5EIX - PubMed Abstract:
Klebsiella pneumoniae is a Gram-negative bacterium that is responsible for a range of common infections, including pulmonary pneumonia, bloodstream infections and meningitis. Certain strains of Klebsiella have become highly resistant to antibiotics. Despite the vast amount of research carried out on this class of bacteria, the molecular structure of its topoisomerase IV, a type II topoisomerase essential for catalysing chromosomal segregation, had remained unknown. In this paper, the structure of its DNA-cleavage complex is reported at 3.35 Å resolution. The complex is comprised of ParC breakage-reunion and ParE TOPRIM domains of K. pneumoniae topoisomerase IV with DNA stabilized by levofloxacin, a broad-spectrum fluoroquinolone antimicrobial agent. This complex is compared with a similar complex from Streptococcus pneumoniae, which has recently been solved.
Organizational Affiliation:
Randall Division of Cell and Molecular Biophysics, King's College London, 3rd Floor, New Hunt's House, Guy's Campus, London SE1 1UL, England.