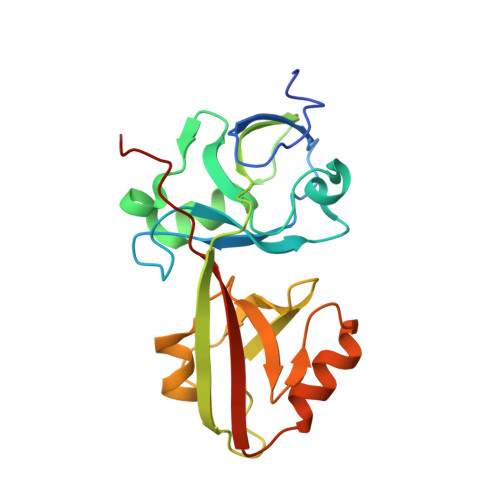
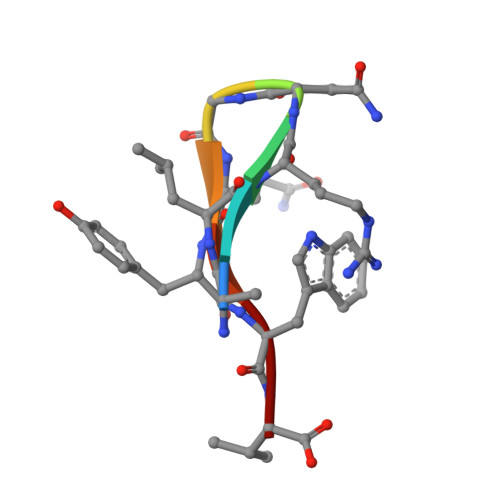
The INAD scaffold is a dynamic, redox-regulated modulator of signaling in the Drosophila eye
Liu, W., Wen, W., Wei, Z., Yu, J., Ye, F., Liu, C.-H., Hardie, R.C., Zhang, M.(2011) Cell 145: 1088-1101
- PubMed: 21703451
- DOI: https://doi.org/10.1016/j.cell.2011.05.015
- Primary Citation of Related Structures:
2LA8, 3R0H - PubMed Abstract:
INAD is a scaffolding protein that regulates signaling in Drosophila photoreceptors. One of its PDZ domains, PDZ5, cycles between reduced and oxidized forms in response to light, but it is unclear how light affects its redox potential. Through biochemical and structural studies, we show that the redox potential of PDZ5 is allosterically regulated by its interaction with another INAD domain, PDZ4. Whereas isolated PDZ5 is stable in the oxidized state, formation of a PDZ45 "supramodule" locks PDZ5 in the reduced state by raising the redox potential of its Cys606/Cys645 disulfide bond by ∼330 mV. Acidification, potentially mediated via light and PLCβ-mediated hydrolysis of PIP(2), disrupts the interaction between PDZ4 and PDZ5, leading to PDZ5 oxidation and dissociation from the TRP Ca(2+) channel, a key component of fly visual signaling. These results show that scaffolding proteins can actively modulate the intrinsic redox potentials of their disulfide bonds to exert regulatory roles in signaling.
Organizational Affiliation:
Division of Life Science, Molecular Neuroscience Center, State Key Laboratory of Molecular Neuroscience, Hong Kong University of Science and Technology, Clear Water Bay, Kowloon, Hong Kong.