Structure of CRISPR-associated protein Csn2
Ellinger, P., Arslan, Z., Wurm, R., Tschapek, B., Pfeffer, K., Wagner, R., Schmitt, L., Pul, U., Smits, S.H.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Sag0897 family CRISPR-associated protein | 229 | Streptococcus agalactiae ATCC 13813 | Mutation(s): 0 Gene Names: HMPREF9171_1216 EC: 2.7.7.6 | 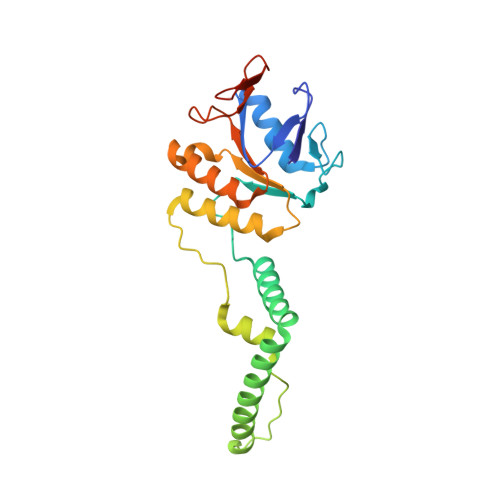 | |
UniProt | |||||
Find proteins for E7S4M0 (Streptococcus agalactiae (strain ATCC 13813 / DSM 2134 / JCM 5671 / NCIMB 701348 / NCTC 8181)) Explore E7S4M0 Go to UniProtKB: E7S4M0 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | E7S4M0 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| EDO Query on EDO | E [auth A], I [auth B], J [auth B] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| CA Query on CA | C [auth A], D [auth A], F [auth B], G [auth B], H [auth B] | CALCIUM ION Ca BHPQYMZQTOCNFJ-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 75.3 | α = 90 |
| b = 83.3 | β = 109.4 |
| c = 110.4 | γ = 90 |
| Software Name | Purpose |
|---|---|
| BEST | data collection |
| Auto-Rickshaw | phasing |
| REFMAC | refinement |
| XDS | data reduction |
| XSCALE | data scaling |