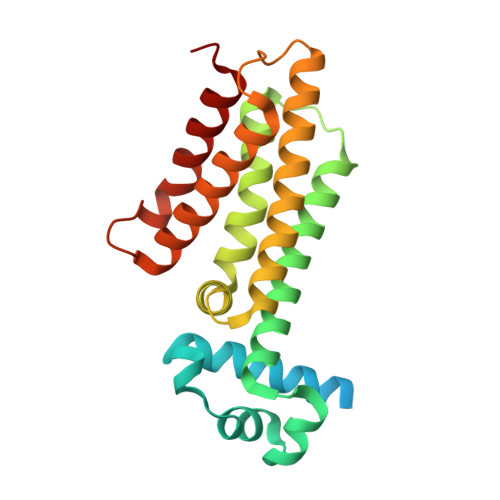
Design, synthesis and optimization of new EthR inhibitors. A new alternative approach to fight tuberculosis by boosting ethionamide
Flipo, M., Desroses, M., Dirie, B., Carette, X., Lens, Z., Rucktooa, P., Leroux, F., Piveteau, C., Demirkaya, F., Locht, C., Villeret, V., Christophe, T., Jeon, H.K., Brodin, P., Deprez, B., Baulard, A., Willand, N.To be published.