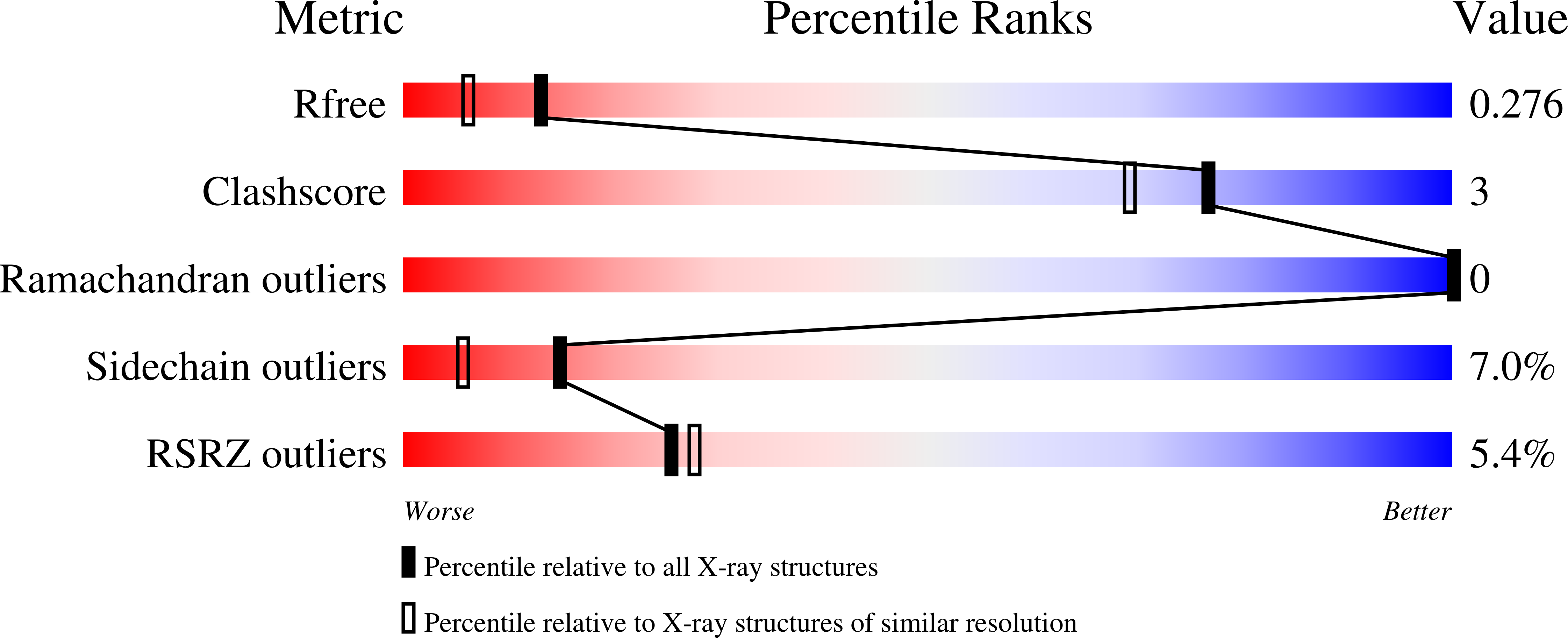
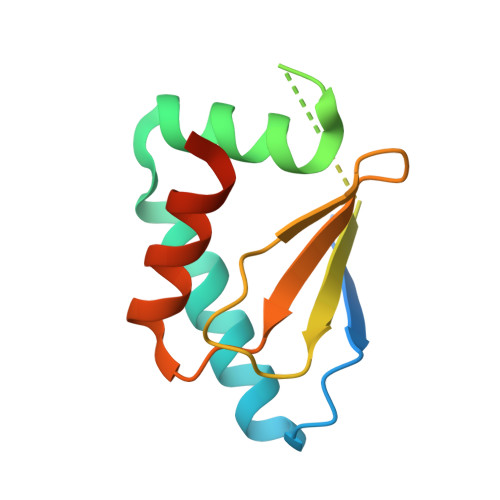
Structure of the HECT C-lobe of the UBR5 E3 ubiquitin ligase.
Matta-Camacho, E., Kozlov, G., Menade, M., Gehring, K.(2012) Acta Crystallogr Sect F Struct Biol Cryst Commun 68: 1158-1163
- PubMed: 23027739
- DOI: https://doi.org/10.1107/S1744309112036937
- Primary Citation of Related Structures:
3PT3 - PubMed Abstract:
UBR5 ubiquitin ligase (also known as EDD, Rat100 or hHYD) is a member of the E3 protein family of HECT (homologous to E6-AP C-terminus) ligases as it contains a C-terminal HECT domain. In ubiquitination cascades involving E3s of the HECT class, ubiquitin is transferred from an associated E2 ubiquitin-conjugating enzyme to the acceptor cysteine of the HECT domain, which consists of structurally distinct N- and C-lobes connected by a flexible linker. Here, the high-resolution crystal structure of the C-lobe of the HECT domain of human UBR5 is presented. The structure reveals important features that are unique compared with other HECT domains. In particular, a distinct four-residue insert in the second helix elongates this helix, resulting in a strikingly different orientation of the preceding loop. This protruding loop is likely to contribute to specificity towards the E2 ubiquitin-conjugating enzyme UBCH4, which is an important functional partner of UBR5. Ubiquitination assays showed that the C-lobe of UBR5 is able to form a thioester-linked E3-ubiquitin complex, although it does not physically interact with UBCH4 in NMR experiments. This study contributes to a better understanding of UBR5 ubiquitination activity.
Organizational Affiliation:
Groupe de Recherche axé sur la Structure des Protéines, Department of Biochemistry, McGill University, Montreal, Quebec H3G 0B1, Canada.