Importance of single molecular determinants in the fidelity of expanded genetic codes.
Antonczak, A.K., Simova, Z., Yonemoto, I.T., Bochtler, M., Piasecka, A., Czapinska, H., Brancale, A., Tippmann, E.M.(2011) Proc Natl Acad Sci U S A 108: 1320-1325
- PubMed: 21224416
- DOI: https://doi.org/10.1073/pnas.1012276108
- Primary Citation of Related Structures:
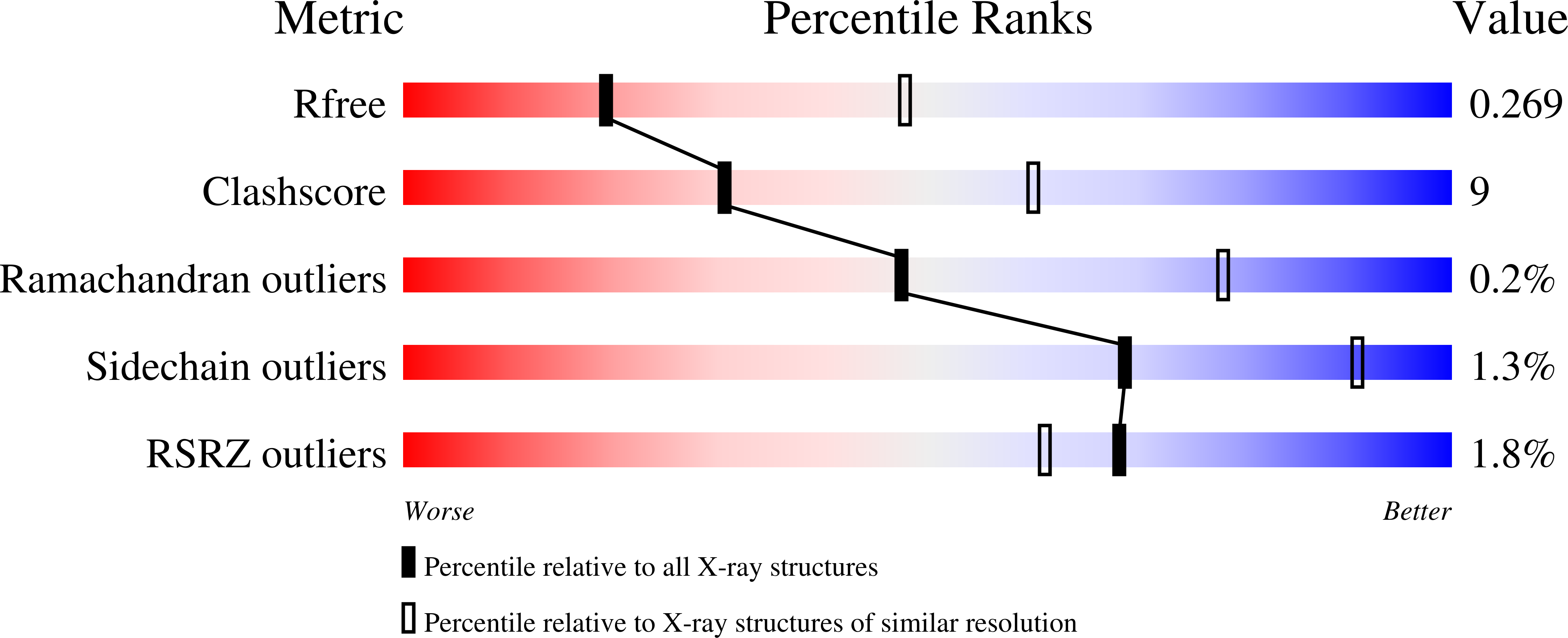
3PRH - PubMed Abstract:
The site-selective encoding of noncanonical amino acids (NAAs) is a powerful technique for the installation of novel chemical functional groups in proteins. This is often achieved by recoding a stop codon and requires two additional components: an evolved aminoacyl tRNA synthetase (AARS) and a cognate tRNA. Analysis of the most successful AARSs reveals common characteristics. The highest fidelity NAA systems derived from the Methanocaldococcus jannaschii tyrosyl AARS feature specific mutations to two residues reported to interact with the hydroxyl group of the substrate tyrosine. We demonstrate that the restoration of just one of these determinants for amino acid specificity results in the loss of fidelity as the evolved AARSs become noticeably promiscuous. These results offer a partial explanation of a recently retracted strategy for the synthesis of glycoproteins. Similarly, we reinvestigated a tryptophanyl AARS reported to allow the site-selective incorporation of 5-hydroxy tryptophan within mammalian cells. In multiple experiments, the enzyme displayed elements of promiscuity despite its previous characterization as a high fidelity enzyme. Given the many similarities of the TyrRSs and TrpRSs reevaluated here, our findings can be largely combined, and in doing so they reinforce the long-established central dogma regarding the molecular basis by which these enzymes contribute to the fidelity of translation. Thus, our view is that the central claims of fidelity reported in several NAA systems remain unproven and unprecedented.
Organizational Affiliation:
School of Chemistry, Cardiff University, Cardiff CF10 3AT United Kingdom.