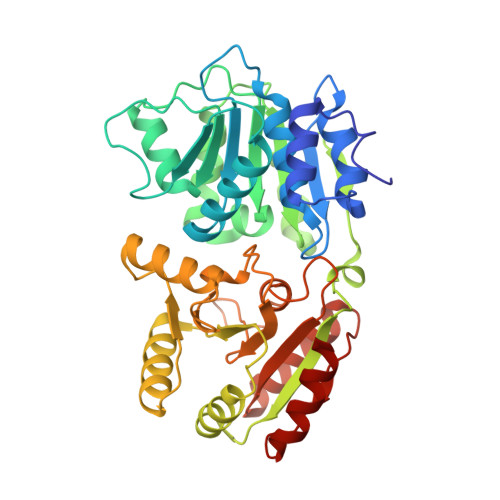
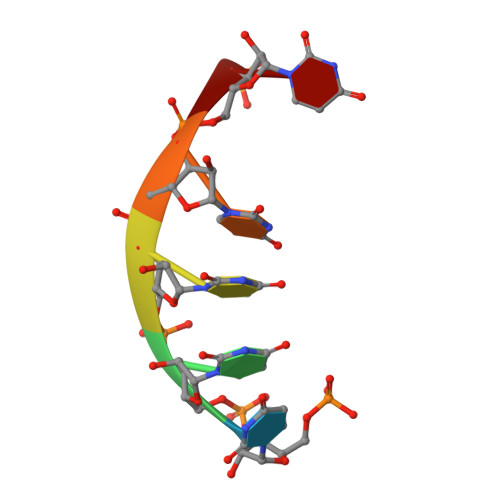
A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP(6) in mRNA export.
Montpetit, B., Thomsen, N.D., Helmke, K.J., Seeliger, M.A., Berger, J.M., Weis, K.(2011) Nature 472: 238-242
- PubMed: 21441902
- DOI: https://doi.org/10.1038/nature09862
- Primary Citation of Related Structures:
3PEU, 3PEV, 3PEW, 3PEY, 3RRM, 3RRN - PubMed Abstract:
Superfamily 1 and superfamily 2 RNA helicases are ubiquitous messenger-RNA-protein complex (mRNP) remodelling enzymes that have critical roles in all aspects of RNA metabolism. The superfamily 2 DEAD-box ATPase Dbp5 (human DDX19) functions in mRNA export and is thought to remodel mRNPs at the nuclear pore complex (NPC). Dbp5 is localized to the NPC via an interaction with Nup159 (NUP214 in vertebrates) and is locally activated there by Gle1 together with the small-molecule inositol hexakisphosphate (InsP(6)). Local activation of Dbp5 at the NPC by Gle1 is essential for mRNA export in vivo; however, the mechanistic role of Dbp5 in mRNP export is poorly understood and it is not known how Gle1(InsP6) and Nup159 regulate the activity of Dbp5. Here we report, from yeast, structures of Dbp5 in complex with Gle1(InsP6), Nup159/Gle1(InsP6) and RNA. These structures reveal that InsP(6) functions as a small-molecule tether for the Gle1-Dbp5 interaction. Surprisingly, the Gle1(InsP6)-Dbp5 complex is structurally similar to another DEAD-box ATPase complex essential for translation initiation, eIF4G-eIF4A, and we demonstrate that Gle1(InsP6) and eIF4G both activate their DEAD-box partner by stimulating RNA release. Furthermore, Gle1(InsP6) relieves Dbp5 autoregulation and cooperates with Nup159 in stabilizing an open Dbp5 intermediate that precludes RNA binding. These findings explain how Gle1(InsP6), Nup159 and Dbp5 collaborate in mRNA export and provide a general mechanism for DEAD-box ATPase regulation by Gle1/eIF4G-like activators.
Organizational Affiliation:
Division of Cell and Developmental Biology, University of California, Berkeley, California 94720, USA.