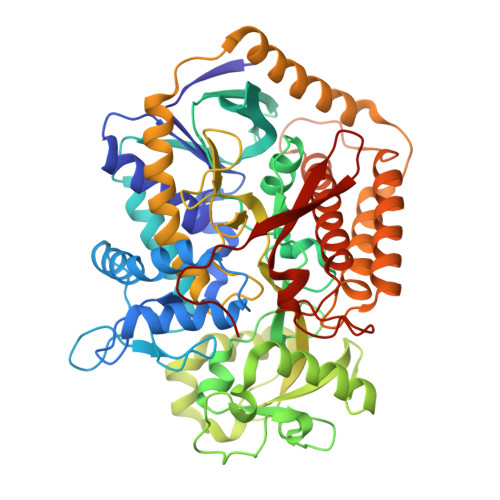
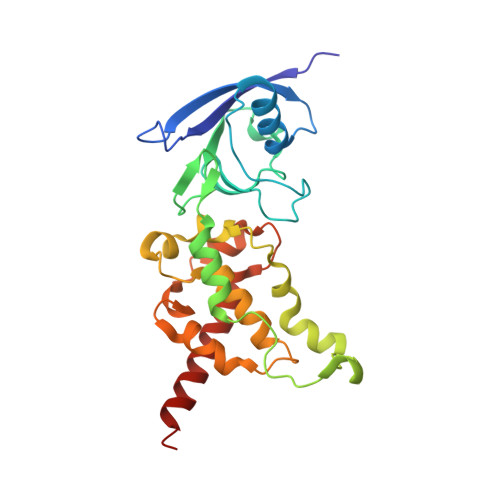
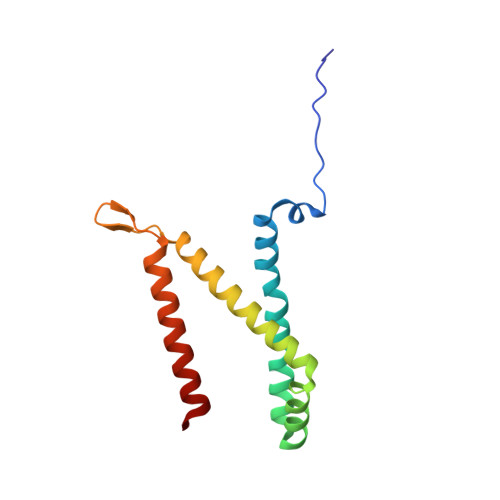
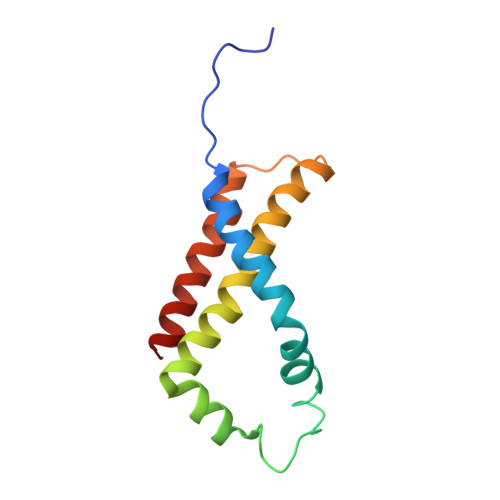
Geometric restraint drives on- and off-pathway catalysis by the Escherichia coli menaquinol:fumarate reductase.
Tomasiak, T.M., Archuleta, T.L., Andrell, J., Luna-Chavez, C., Davis, T.A., Sarwar, M., Ham, A.J., McDonald, W.H., Yankovskaya, V., Stern, H.A., Johnston, J.N., Maklashina, E., Cecchini, G., Iverson, T.M.(2011) J Biol Chem 286: 3047-3056
- PubMed: 21098488
- DOI: https://doi.org/10.1074/jbc.M110.192849
- Primary Citation of Related Structures:
3P4P, 3P4Q, 3P4R, 3P4S - PubMed Abstract:
Complex II superfamily members catalyze the kinetically difficult interconversion of succinate and fumarate. Due to the relative simplicity of complex II substrates and their similarity to other biologically abundant small molecules, substrate specificity presents a challenge in this system. In order to identify determinants for on-pathway catalysis, off-pathway catalysis, and enzyme inhibition, crystal structures of Escherichia coli menaquinol:fumarate reductase (QFR), a complex II superfamily member, were determined bound to the substrate, fumarate, and the inhibitors oxaloacetate, glutarate, and 3-nitropropionate. Optical difference spectroscopy and computational modeling support a model where QFR twists the dicarboxylate, activating it for catalysis. Orientation of the C2-C3 double bond of activated fumarate parallel to the C(4a)-N5 bond of FAD allows orbital overlap between the substrate and the cofactor, priming the substrate for nucleophilic attack. Off-pathway catalysis, such as the conversion of malate to oxaloacetate or the activation of the toxin 3-nitropropionate may occur when inhibitors bind with a similarly activated bond in the same position. Conversely, inhibitors that do not orient an activatable bond in this manner, such as glutarate and citrate, are excluded from catalysis and act as inhibitors of substrate binding. These results support a model where electronic interactions via geometric constraint and orbital steering underlie catalysis by QFR.
Organizational Affiliation:
Department of Pharmacology, Vanderbilt University Medical Center, Nashville, Tennessee 37232, USA.