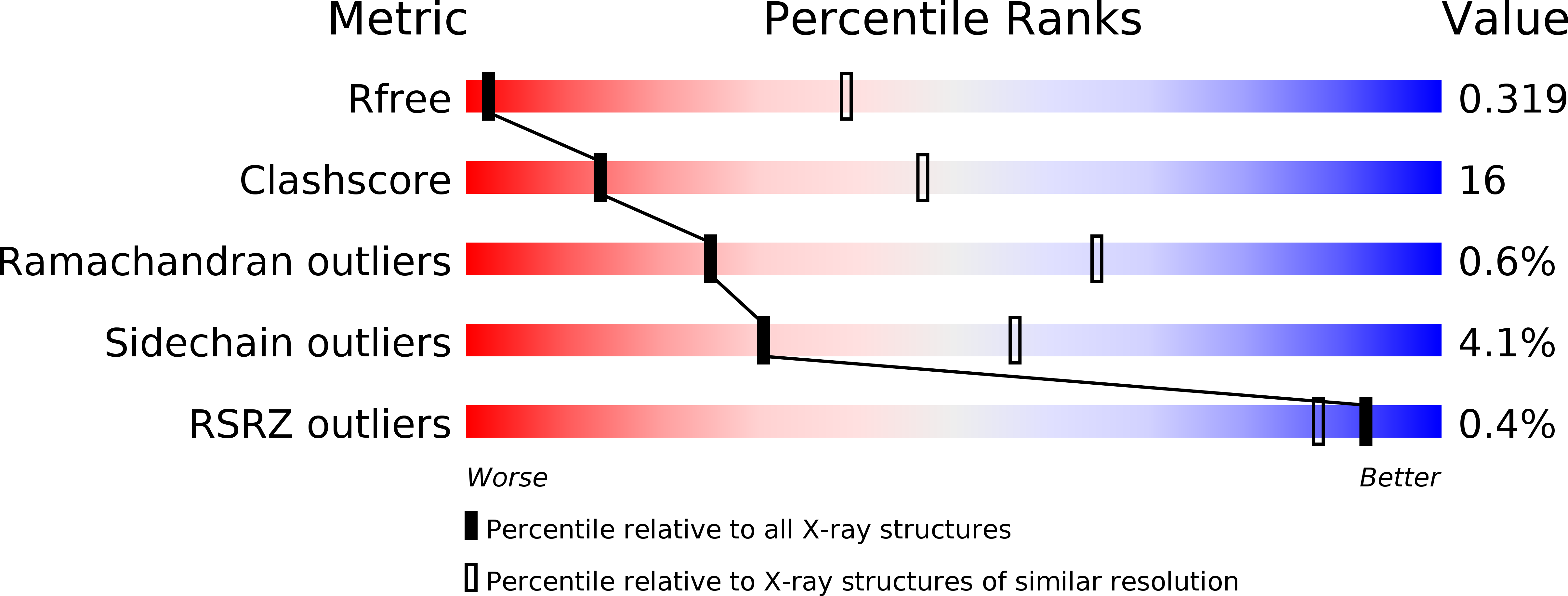
Dynamics and allosteric potential of the AMPA receptor N-terminal domain
Sukumaran, M., Rossmann, M., Shrivastava, I., Dutta, A., Bahar, I., Greger, I.H.(2011) EMBO J 30: 972-982
- PubMed: 21317871
- DOI: https://doi.org/10.1038/emboj.2011.17
- Primary Citation of Related Structures:
3O21, 3P3W - PubMed Abstract:
Glutamate-gated ion channels (ionotropic glutamate receptors, iGluRs) sense the extracellular milieu via an extensive extracellular portion, comprised of two clamshell-shaped segments. The distal, N-terminal domain (NTD) has allosteric potential in NMDA-type iGluRs, which has not been ascribed to the analogous domain in AMPA receptors (AMPARs). In this study, we present new structural data uncovering dynamic properties of the GluA2 and GluA3 AMPAR NTDs. GluA3 features a zipped-open dimer interface with unconstrained lower clamshell lobes, reminiscent of metabotropic GluRs (mGluRs). The resulting labile interface supports interprotomer rotations, which can be transmitted to downstream receptor segments. Normal mode analysis reveals two dominant mechanisms of AMPAR NTD motion: intraprotomer clamshell motions and interprotomer counter-rotations, as well as accessible interconversion between AMPAR and mGluR conformations. In addition, we detect electron density for a potential ligand in the GluA2 interlobe cleft, which may trigger lobe motions. Together, these data support a dynamic role for the AMPAR NTDs, which widens the allosteric landscape of the receptor and could provide a novel target for ligand development.
Organizational Affiliation:
Neurobiology Division, MRC Laboratory of Molecular Biology, Cambridge, UK.