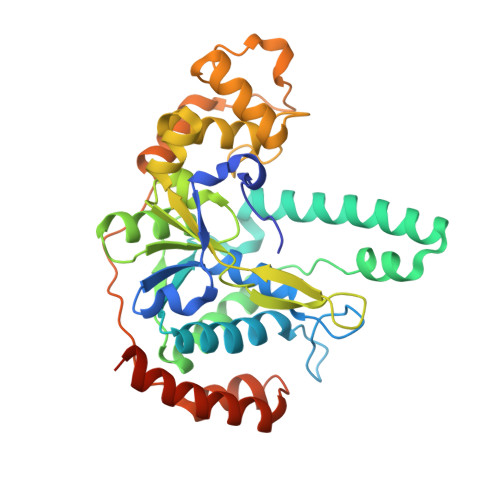
Structure of flap endonuclease 1 from the hyperthermophilic archaeon Desulfurococcus amylolyticus
Mase, T., Kubota, K., Miyazono, K., Kawarabayasi, Y., Tanokura, M.(2011) Acta Crystallogr Sect F Struct Biol Cryst Commun 67: 209-213
- PubMed: 21301087
- DOI: https://doi.org/10.1107/S1744309110053030
- Primary Citation of Related Structures:
3ORY - PubMed Abstract:
Flap endonuclease 1 (FEN1) is a key enzyme in DNA repair and DNA replication. It is a structure-specific nuclease that removes 5'-overhanging flaps and the RNA/DNA primer during maturation of the Okazaki fragment. Homologues of FEN1 exist in a wide range of bacteria, archaea and eukaryotes. In order to further understand the structural basis of the DNA recognition, binding and cleavage mechanism of FEN1, the structure of FEN1 from the hyperthermophilic archaeon Desulfurococcus amylolyticus (DaFEN1) was determined at 2.00 Å resolution. The overall fold of DaFEN1 was similar to those of other archaeal FEN1 proteins; however, the helical clamp and the flexible loop exhibited a putative substrate-binding pocket with a unique conformation.
Organizational Affiliation:
Department of Applied Biological Chemistry, Graduate School of Agricultural and Life Sciences, The University of Tokyo, Tokyo, Japan.