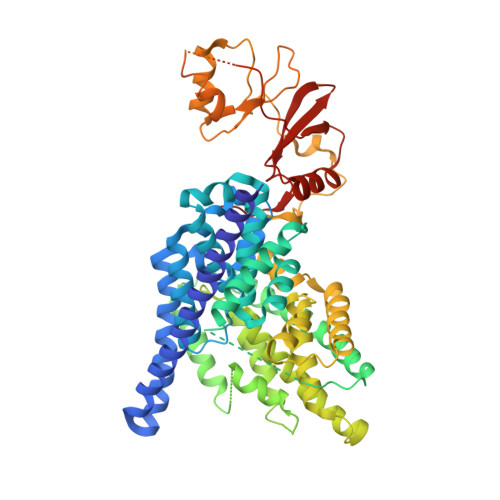
Structure of a eukaryotic CLC transporter defines an intermediate state in the transport cycle.
Feng, L., Campbell, E.B., Hsiung, Y., MacKinnon, R.(2010) Science 330: 635-641
- PubMed: 20929736
- DOI: https://doi.org/10.1126/science.1195230
- Primary Citation of Related Structures:
3ORG - PubMed Abstract:
CLC proteins transport chloride (Cl(-)) ions across cell membranes to control the electrical potential of muscle cells, transfer electrolytes across epithelia, and control the pH and electrolyte composition of intracellular organelles. Some members of this protein family are Cl(-) ion channels, whereas others are secondary active transporters that exchange Cl(-) ions and protons (H(+)) with a 2:1 stoichiometry. We have determined the structure of a eukaryotic CLC transporter at 3.5 angstrom resolution. Cytoplasmic cystathionine beta-synthase (CBS) domains are strategically positioned to regulate the ion-transport pathway, and many disease-causing mutations in human CLCs reside on the CBS-transmembrane interface. Comparison with prokaryotic CLC shows that a gating glutamate residue changes conformation and suggests a basis for 2:1 Cl(-)/H(+) exchange and a simple mechanistic connection between CLC channels and transporters.
Organizational Affiliation:
Laboratory of Molecular Neurobiology and Biophysics, Rockefeller University, Howard Hughes Medical Institute, 1230 York Avenue, New York, NY 10065, USA.