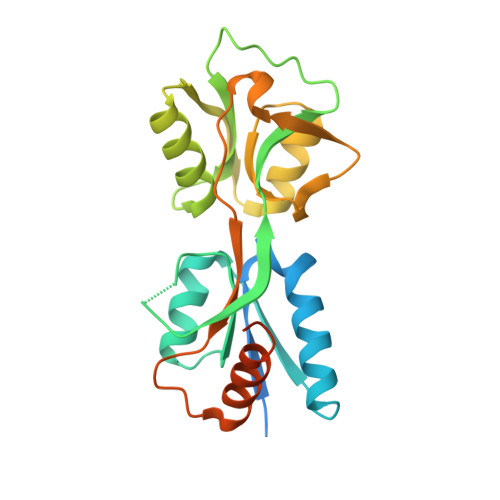
Structure of the effector-binding domain of the LysR-type transcription factor RovM from Yersinia pseudotuberculosis.
Quade, N., Dieckmann, M., Haffke, M., Heroven, A.K., Dersch, P., Heinz, D.W.(2011) Acta Crystallogr D Biol Crystallogr 67: 81-90
- PubMed: 21245528
- DOI: https://doi.org/10.1107/S0907444910049681
- Primary Citation of Related Structures:
3ONM - PubMed Abstract:
In enteropathogenic Yersinia, the expression of several early-phase virulence factors such as invasin is tightly regulated in response to environmental cues. The responsible regulatory network is complex, involving several regulatory RNAs and proteins such as the LysR-type transcription regulator (LTTR) RovM. In this study, the crystal structure of the effector-binding domain (EBD) of RovM, the first LTTR protein described as being involved in virulence regulation, was determined at a resolution of 2.4 Å. Size-exclusion chromatography and comparison with structures of full-length LTTRs show that RovM is most likely to adopt a tetrameric arrangement with two distant DNA-binding domains (DBDs), causing the DNA to bend around it. Additionally, a cavity was detected in RovM which could bind small inducer molecules.
Organizational Affiliation:
Department of Molecular Structural Biology, Helmholtz Centre for Infection Research, D-38124 Braunschweig, Germany.