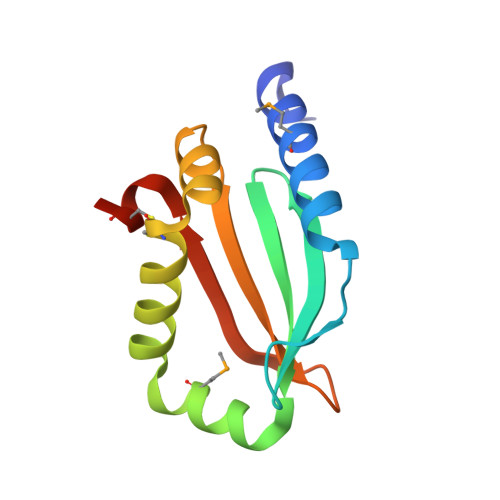
Crystal structure of a putative quorum sensing-regulated protein (PA3611) from the Pseudomonas-specific DUF4146 family.
Das, D., Chiu, H.J., Farr, C.L., Grant, J.C., Jaroszewski, L., Knuth, M.W., Miller, M.D., Tien, H.J., Elsliger, M.A., Deacon, A.M., Godzik, A., Lesley, S.A., Wilson, I.A.(2014) Proteins 82: 1086-1092
- PubMed: 24174223
- DOI: https://doi.org/10.1002/prot.24455
- Primary Citation of Related Structures:
3NPD - PubMed Abstract:
Pseudomonas aeruginosa is an opportunistic pathogen commonly found in humans and other organisms and is an important cause of infection especially in patients with compromised immune defense mechanisms. The PA3611 gene of P. aeruginosa PAO1 encodes a secreted protein of unknown function, which has been recently classified into a small Pseudomonas-specific protein family called DUF4146. As part of our effort to extend structural coverage of novel protein space and provide a structure-based functional insight into new protein families, we report the crystal structure of PA3611, the first structural representative of the DUF4146 protein family.
Organizational Affiliation:
Joint Center for Structural Genomics; Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, Menlo Park, California.