Determination of the X-ray structure of the snake venom protein omwaprin by total chemical synthesis and racemic protein crystallography.
Banigan, J.R., Mandal, K., Sawaya, M.R., Thammavongsa, V., Hendrickx, A.P., Schneewind, O., Yeates, T.O., Kent, S.B.(2010) Protein Sci 19: 1840-1849
- PubMed: 20669184
- DOI: https://doi.org/10.1002/pro.468
- Primary Citation of Related Structures:
3NGG - PubMed Abstract:
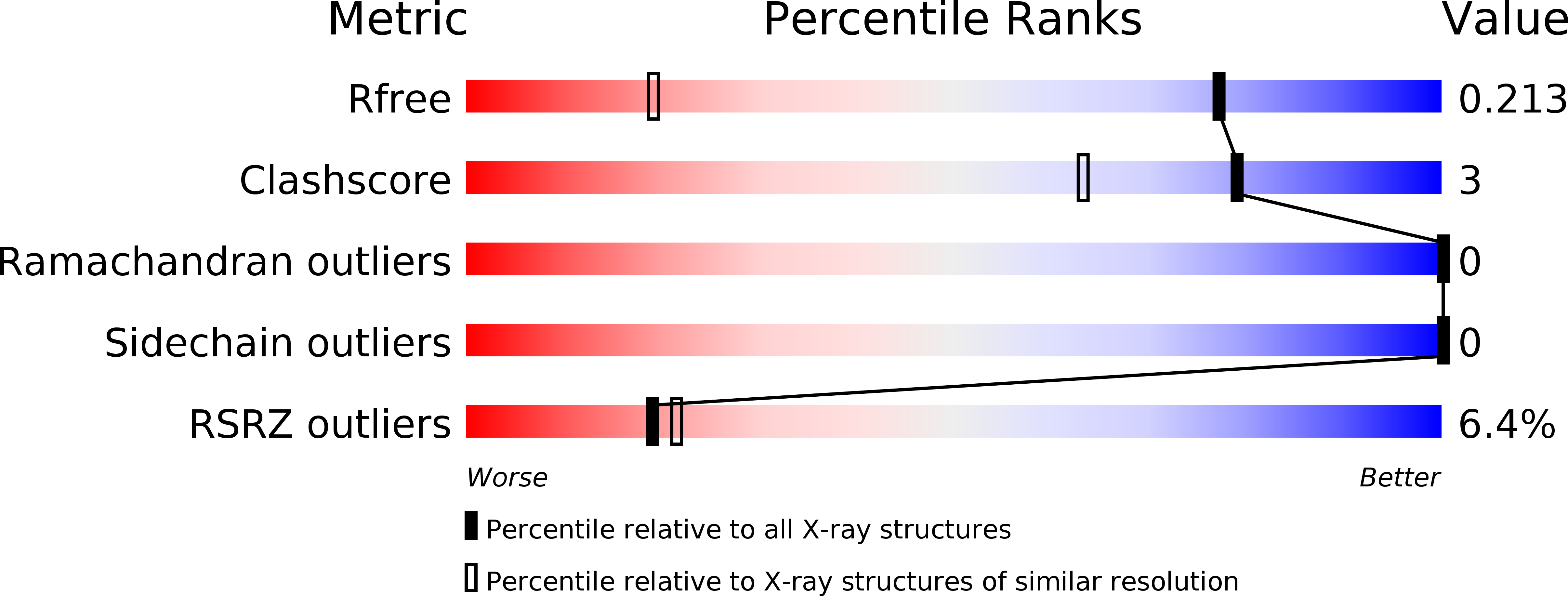
The 50-residue snake venom protein L-omwaprin and its enantiomer D-omwaprin were prepared by total chemical synthesis. Radial diffusion assays were performed against Bacillus megaterium and Bacillus anthracis; both L- and D-omwaprin showed antibacterial activity against B. megaterium. The native protein enantiomer, made of L-amino acids, failed to crystallize readily. However, when a racemic mixture containing equal amounts of L- and D-omwaprin was used, diffraction quality crystals were obtained. The racemic protein sample crystallized in the centrosymmetric space group P2(1)/c and its structure was determined at atomic resolution (1.33 A) by a combination of Patterson and direct methods based on the strong scattering from the sulfur atoms in the eight cysteine residues per protein. Racemic crystallography once again proved to be a valuable method for obtaining crystals of recalcitrant proteins and for determining high-resolution X-ray structures by direct methods.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, The University of Chicago, Chicago, Illinois, USA.