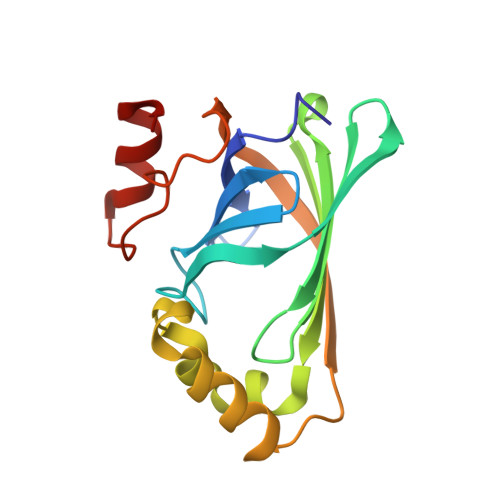
Structural analysis of Bacillus pumilus phenolic acid decarboxylase, a lipocalin-fold enzyme.
Matte, A., Grosse, S., Bergeron, H., Abokitse, K., Lau, P.C.(2010) Acta Crystallogr Sect F Struct Biol Cryst Commun 66: 1407-1414
- PubMed: 21045284
- DOI: https://doi.org/10.1107/S174430911003246X
- Primary Citation of Related Structures:
3NAD - PubMed Abstract:
The decarboxylation of phenolic acids, including ferulic and p-coumaric acids, to their corresponding vinyl derivatives is of importance in the flavouring and polymer industries. Here, the crystal structure of phenolic acid decarboxylase (PAD) from Bacillus pumilus strain UI-670 is reported. The enzyme is a 161-residue polypeptide that forms dimers both in the crystal and in solution. The structure of PAD as determined by X-ray crystallography revealed a β-barrel structure and two α-helices, with a cleft formed at one edge of the barrel. The PAD structure resembles those of the lipocalin-fold proteins, which often bind hydrophobic ligands. Superposition of structurally related proteins bound to their cognate ligands shows that they and PAD bind their ligands in a conserved location within the β-barrel. Analysis of the residue-conservation pattern for PAD-related sequences mapped onto the PAD structure reveals that the conservation mainly includes residues found within the hydrophobic core of the protein, defining a common lipocalin-like fold for this enzyme family. A narrow cleft containing several conserved amino acids was observed as a structural feature and a potential ligand-binding site.
Organizational Affiliation:
Health Sector, Biotechnology Research Institute, 6100 Royalmount Avenue, Montreal, Quebec H4P 2R2, Canada.