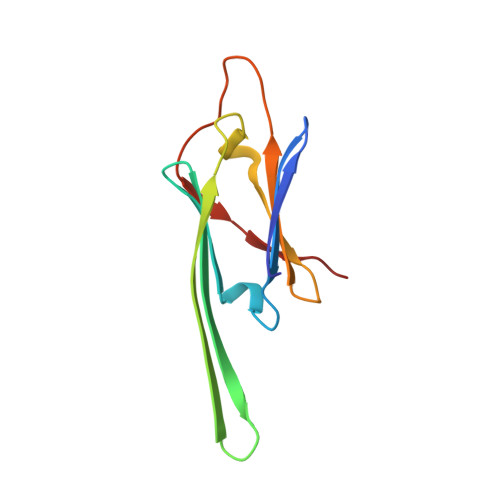
Non-3D domain swapped crystal structure of truncated zebrafish alphaA crystallin.
Laganowsky, A., Eisenberg, D.(2010) Protein Sci 19: 1978-1984
- PubMed: 20669149
- DOI: https://doi.org/10.1002/pro.471
- Primary Citation of Related Structures:
3N3E - PubMed Abstract:
In previous work on truncated alpha crystallins (Laganowsky et al., Protein Sci 2010; 19:1031-1043), we determined crystal structures of the alpha crystallin core, a seven beta-stranded immunoglobulin-like domain, with its conserved C-terminal extension. These extensions swap into neighboring cores forming oligomeric assemblies. The extension is palindromic in sequence, binding in either of two directions. Here, we report the crystal structure of a truncated alphaA crystallin (AAC) from zebrafish (Danio rerio) revealing C-terminal extensions in a non three-dimensional (3D) domain swapped, "closed" state. The extension is quasi-palindromic, bound within its own zebrafish core domain, lying in the opposite direction to that of bovine AAC, which is bound within an adjacent core domain (Laganowsky et al., Protein Sci 2010; 19:1031-1043). Our findings establish that the C-terminal extension of alpha crystallin proteins can be either 3D domain swapped or non-3D domain swapped. This duality provides another molecular mechanism for alpha crystallin proteins to maintain the polydispersity that is crucial for eye lens transparency.
Organizational Affiliation:
UCLA-DOE, Institute for Genomics and Proteomics, Los Angeles, California 90095-1570, USA.