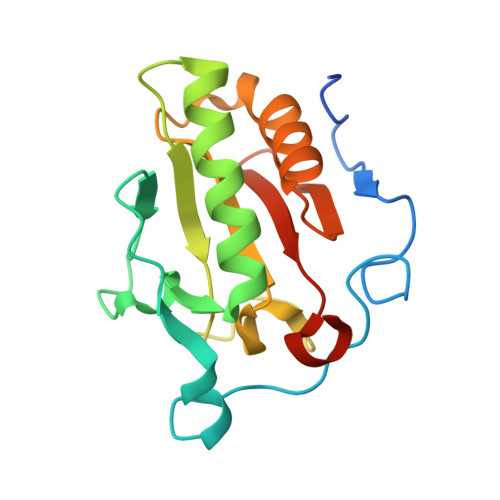
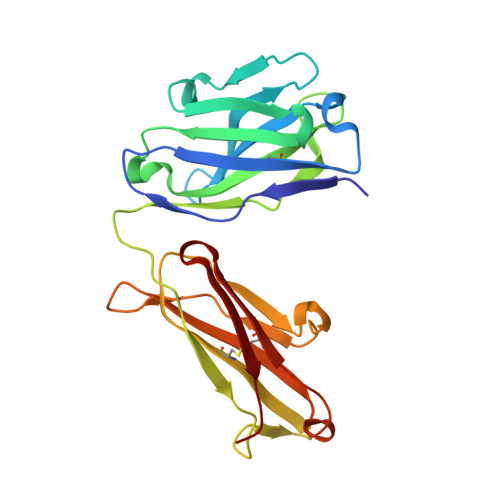
Hedgehog pathway antagonist 5E1 binds hedgehog at the pseudo-active site.
Maun, H.R., Wen, X., Lingel, A., de Sauvage, F.J., Lazarus, R.A., Scales, S.J., Hymowitz, S.G.(2010) J Biol Chem 285: 26570-26580
- PubMed: 20504762
- DOI: https://doi.org/10.1074/jbc.M110.112284
- Primary Citation of Related Structures:
3MXV, 3MXW - PubMed Abstract:
Proper hedgehog (Hh) signaling is crucial for embryogenesis and tissue regeneration. Dysregulation of this pathway is associated with several types of cancer. The monoclonal antibody 5E1 is a Hh pathway inhibitor that has been extensively used to elucidate vertebrate Hh biology due to its ability to block binding of the three mammalian Hh homologs to the receptor, Patched1 (Ptc1). Here, we engineered a murine:human chimeric 5E1 (ch5E1) with similar Hh-binding properties to the original murine antibody. Using biochemical, biophysical, and x-ray crystallographic studies, we show that, like the regulatory receptors Cdon and Hedgehog-interacting protein (Hhip), ch5E1 binding to Sonic hedgehog (Shh) is enhanced by calcium ions. In the presence of calcium and zinc ions, the ch5E1 binding affinity increases 10-20-fold to tighter than 1 nm primarily because of a decrease in the dissociation rate. The co-crystal structure of Shh bound to the Fab fragment of ch5E1 reveals that 5E1 binds at the pseudo-active site groove of Shh with an epitope that largely overlaps with the binding site of its natural receptor antagonist Hhip. Unlike Hhip, the side chains of 5E1 do not directly coordinate the Zn(2+) cation in the pseudo-active site, despite the modest zinc-dependent increase in 5E1 affinity for Shh. Furthermore, to our knowledge, the ch5E1 Fab-Shh complex represents the first structure of an inhibitor antibody bound to a metalloprotease fold.
Organizational Affiliation:
Department of Protein Engineering, Genentech, Inc., South San Francisco, California 94080, USA.