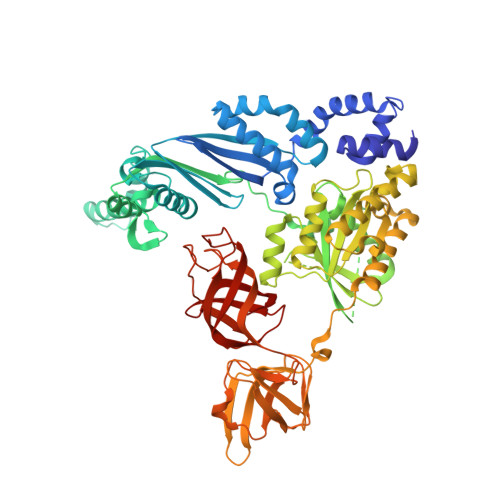
Structure of the Qbeta replicase, an RNA-dependent RNA polymerase consisting of viral and host proteins.
Kidmose, R.T., Vasiliev, N.N., Chetverin, A.B., Andersen, G.R., Knudsen, C.R.(2010) Proc Natl Acad Sci U S A 107: 10884-10889
- PubMed: 20534494
- DOI: https://doi.org/10.1073/pnas.1003015107
- Primary Citation of Related Structures:
3MMP - PubMed Abstract:
The RNA-dependent RNA polymerase core complex formed upon infection of Escherichia coli by the bacteriophage Qbeta is composed of the viral catalytic beta-subunit as well as the host translation elongation factors EF-Tu and EF-Ts, which are required for initiation of RNA replication. We have determined the crystal structure of the complex between the beta-subunit and the two host proteins to 2.5-A resolution. Whereas the basic catalytic machinery in the viral subunit appears similar to other RNA-dependent RNA polymerases, a unique C-terminal region of the beta-subunit engages in extensive interactions with EF-Tu and may contribute to the separation of the transient duplex formed between the template and the nascent product to allow exponential amplification of the phage genome. The evolution of resistance by the host appears to be impaired because of the interactions of the beta-subunit with parts of EF-Tu essential in recognition of aminoacyl-tRNA.
Organizational Affiliation:
Department of Molecular Biology, University of Aarhus, Gustav Wieds Vej 10C, DK-8000 Aarhus, Denmark.