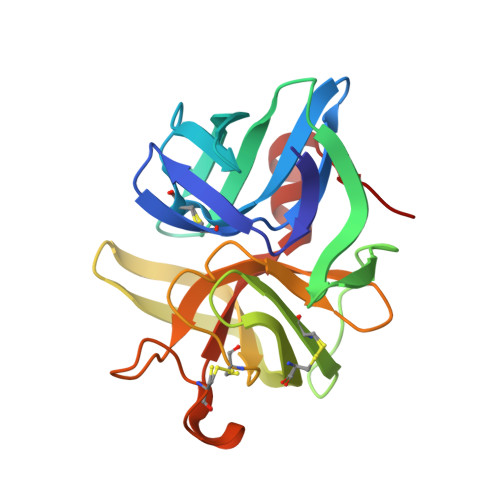
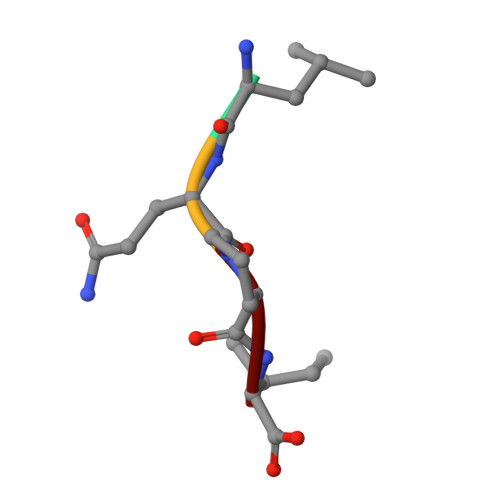
Quantifying protein unfolding cooperativity with acid sensitive probes: Interdomain salt bridge contributions to unfolding cooperativity are combined efficiently in alpha-Lytic Protease
Erciyas Bailey, F.P., Waddling, C.A., Agard, D.A.To be published.