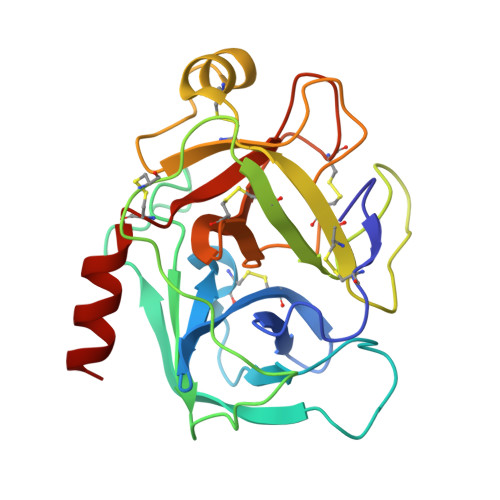
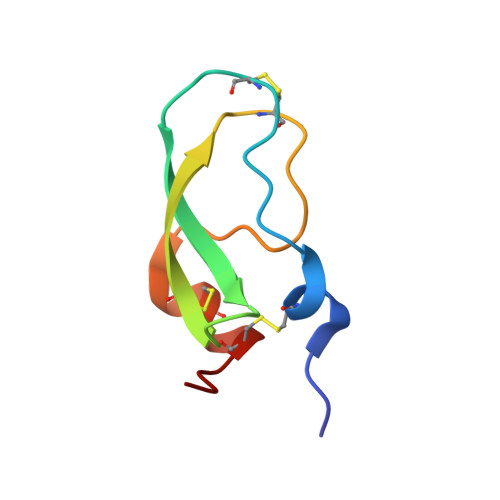
Structural insights into serine protease inhibition by a marine invertebrate BPTI Kunitz-type inhibitor.
Garcia-Fernandez, R., Pons, T., Perbandt, M., Valiente, P.A., Talavera, A., Gonzalez-Gonzalez, Y., Rehders, D., Chavez, M.A., Betzel, C., Redecke, L.(2012) J Struct Biol 180: 271-279
- PubMed: 22975140
- DOI: https://doi.org/10.1016/j.jsb.2012.08.009
- Primary Citation of Related Structures:
3M7Q - PubMed Abstract:
Proteins isolated from marine invertebrates are frequently characterized by exceptional structural and functional properties. ShPI-1, a BPTI Kunitz-type inhibitor from the Caribbean Sea anemone Stichodactyla helianthus, displays activity not only against serine-, but also against cysteine-, and aspartate proteases. As an initial step to evaluate the molecular basis of its activities, we describe the crystallographic structure of ShPI-1 in complex with the serine protease bovine pancreatic trypsin at 1.7Å resolution. The overall structure and the important enzyme-inhibitor interactions of this first invertebrate BPTI-like Kunitz-type inhibitor:trypsin complex remained largely conserved compared to mammalian BPTI-Kunitz inhibitor complexes. However, a prominent stabilizing role within the interface was attributed to arginine at position P3. Binding free-energy calculations indicated a 10-fold decrease for the inhibitor affinity against trypsin, if the P3 residue of ShPI-1 is mutated to alanine. Together with the increased role of Arg(11) at P3 position, slightly reduced interactions at the prime side (Pn') of the primary binding loop and at the secondary binding loop of ShPI-1 were detected. In addition, the structure provides important information for site directed mutagenesis to further optimize the activity of rShPI-1A for biotechnological applications.
Organizational Affiliation:
Centro de Estudio de Proteínas, Facultad de Biología, Universidad de la Habana, Calle 25 No 411, Havana, Cuba.